MOJ
eISSN: 2374-6920


Research Article Volume 9 Issue 2
1Virology department, Pasteur Institute of Iran, Tehran, Iran
2Microbiology department, Pasteur Institute of Iran, Tehran, Iran
3Biophysics Department, Faculty of Biological Sciences, Tarbiat Modares University (TMU), Tehran, Iran
4Nano-biotechnology department, Pasteur Institute of Iran, Tehran, Iran
Correspondence: Seyed Shahriar Arab, Biophysics Department, School of Biological Sciences, Tarbiat Modares University (TMU), Nasr, Jalal AleAhmad, Tehran, Iran
Received: April 20, 2020 | Published: June 29, 2020
Citation: Kazemi F, Arab SS, Roohvand F, et al. Molecular dynamics simulation of SKG132: An initiation to understand the mechanism of plasminogen activation. MOJ Proteomics Bioinform. 2020;9(2):59-63. DOI: 10.15406/mojpb.2020.09.00280
To gain insights into the high plasminogen (Pg) activation of SKG132 from group G (SKG) streptococci, molecular dynamics (MD) simulation was carried out for both standard SK from group C (SKC) and SKG132 with four natural residual substitutions (Ile33Phe, Arg45Gln, Asn228Lys, Phe287Ile). The aim of this study was to examine the structural alterations induced by mentioned substitutions in SKG132. Structural analysis of MD simulation results showed two regions with increased flexibility; residues 156-161 and 183-187 which structurally located around 250 loop(251-264). In addition, residues 301-372 showed decreased flexibility in SKγ of SKG132. Presence of enhanced flexible region around functionally important 250 loop and also formation a stable SK.Pg complex due to decreased flexibility of 301-372 region, may lead to improve substrate processing and high Pg activation of SKG132. Moreover, results of protein interaction calculators (PIC), showed presence of hydrogen bond in residue pairs of Lys256-Lys183, Lys256-Glu272 and Lys257-Glu272 in SKC, while they were disrupted in SKG132 and led to formation of three unbinding crucial residues including Lys256, Lys257 and Glu272. Therefore, it is concluded that better exposure of 250 loop for substrate, presence of three unbinding critical residues (Lys256, Lys257 and Glu272) and formation of a stable complex, probably play important roles in increased Pg activation of SKG132 by participating in effective interactions with substrate plasminogen. These results showed the structural differences between SKG132 and SKC in atomic details. The results may be further used for design of SK drug with altered conformation and enhanced activity.
Keywords: streptokinase, molecular dynamics simulations, structural dynamics, streptokinase activity
Streptokinase, a bacterial thrombolytic agent activates fibrinolytic system indirectly. The mechanism of activation initiates with formation of an equimolar complex with plasminogen (Pg) as a partner (SK.Pg: activator complex).1 Upon formation of activator complex, a series of conformational changes occur in Pg and an enzymatically complex will be generated. The SK.Pg complex recruits free Pg as a substrate and activates it through cleavage of scissile peptide bond in the Arg561-Val562 of Pg substrate (proteolytic activity) and converts it to plasmin.2 The proteolytically generated Pm, can either preferentially replace with Pg in SK.Pg complex, with three times higher affinity for SK compared to that of Pg. Contrary to the SK.Pg, the formed SK.Pm complex display very narrow substrate specificity for conversion of Pg to Pm.1,2 SK is secreted by several beta-hemolytic streptococci (βHS) strains including group A (SKA), C (SKC) and G (SKG) and structurally composed of three domains, SKα (1-146), SKβ (147-290) and SKγ (291-414).3 Experimental studies on different parts of SKβ have shown functionally important properties of this domain for Pg substrate processing.3 Alanine scanning mutagenesis of the core region 230-2904 and deletion of 254-273 region showed putative role of these regions in binding to substrate Pg.5Several experimental studies also implicated the importance of the 250 loop (Ala251-Ile264) in SKβ for Pg substrate recognition and processing.6,7 Deletion of the 250 loop and Ala substitution of Lys256 and Lys257 at the tip of the loop led to reduction in Pg activator activity.6,7 In addition docking approach showed that the Lys256 and Lys257 residues of 250 loop interact with kringle domain 5 (K5) of Pg substrate and facilitate substrate identification.8 Multiple contact sites in SK and Pg are required for binding and formation of a stable activator complex which is prerequisite for substrate recognition and interaction.9 It was also proposed that the C-terminal of SKγ is involved in hydrophobic interactions with Pg partner in activator complex and is crucial for maintaining a stable enzymatic complex to catalyze conversion of substrate Pg to Pm.3,10 Up to now, major attempts are being focused on enhancement of Pg activation of SK with the aim of producing improved thrombolytic drug. Most of the studies were based on experimental deigns and its atomic aspects of Pg activation by SK have been neglected. Therefore, in spite of many experimental studies which have been done until now, the atomic aspects of Pg activation by SK is still a matter of debate.
Kinetic study of recent experimental work has demonstrated high Pg activation for recombinant form of SK from GGSgroup (which called SKG132) compared to standard SK from group C, SKC9542, (here after called SKC).11 Pairwise alignment of protein sequences indicated that SKG132 differs from SKC by four residues including Ile33Phe, Arg45Gln, Asn228Lys and Phe287Ile.Our latest computational study has clarified reasons of enhanced Pg activation by investigation of the interaction between SK and µPm. Although, this study has provided a comparison between form of SKG132.µPm and SKC.µPm; but, atomic level description of SK part is still unclear. So, computational studies for comprehending structural and functional properties of SKG132 is also necessary.Molecular dynamics (MD) simulation is an important computational tool for understanding the macromolecular structure-to-function relationships.12 In present study, first SK molecule hasbeen modeled, then to obtain structures for analyses and gain insights into high Pg activation of SKG132, MD simulation was performed to compare SKC and continued until the systems reached to the equilibrium (85 ns). The simulation results were in excellent accordance with experimental data and let us to provide atomic details into the high Pg activation of SKG132 by observing structural and dynamical elements of the SKC and SKG132 proteins.
Homology modeling of streptokinase
The sequence of the SKC9542,Streptococcus dysgalactiae (S.dysgalactiae subsp. equisimilis), with respective identification code (SKC-2, protein_id: AAC60418.1) was obtained from the NCBI database. To find suitable structures as template, BLAST (Basic Local Alignment Search Tool) web server was used13 against the Protein Data Bank (PDB).14 From the protein BLAST results, PDB codes of 1BML, 1L4Z and 1QQR were selected as template. The models were built using multi-template homology modeling by MODELLER v9.13.15 Reliability of the predicted models were evaluated by the RAMPAGE, validation program via Ramachandran plot assessment.16 To generate contributed mutations, YASARA v.11.11.2 was applied and the target amino acid residues were substituted with desired ones (Ile33Phe, Arg45Gln, Asn228Lys, and Phe287Ile) (Figure 1).17
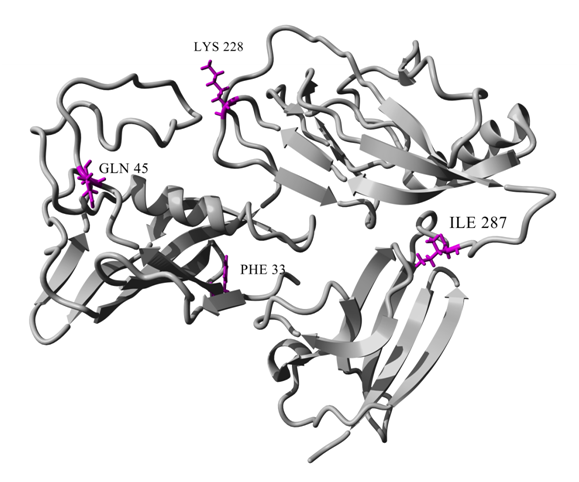
Figure 1 Cartoon representation of modeled SKG132 showing Phe33, Gln45, Lys228 and Ile287 mutations. The color scheme of SKG132 is gray and mutations in SKG132 represented as stick models in dark pink.
Molecular dynamics simulation
The MD simulation of SKC and SKG132 protein structures was performed with the GROMACS v4.6.3 software package.18,19 The GROMOS96 53a6 force field was used for these simulations. The generated models were centered in a dodecahedron box using periodic boundary conditions (PBC). The box was filled with SPC/E water molecules while the protein surface atoms locatedin a minimum spacing of 10 Å from edge of the box. The appropriate numbers of sodium counter ions were added to the box to neutralize the system. Minimization of the system energy was performed using steepest descent and conjugate gradient methods. System equilibration was performed on constant temperature and pressure. To keep the system in equilibrated condition, temperature and pressure were set to 300 K by Berendsenthermostat20 and1 bar using Parrinello-Rahman coupling method, respectively.21 The Particle-Mesh-Ewald (PME) method was used for calculation of electrostatics interactions.22 The covalent bonds length was maintained constant using the Linear Constraint Solver algorithm.23The MD simulation was carried out to equilibrate the protein for 85 ns. Finally, the trajectories of MD simulation were applied for analysis of root mean square deviation (RMSD) [which measures the similarity between two superimposed atomic coordinates], root mean square fluctuation (RMSF) [that measures the flexibility of the Cα atom of each residue] and hydrogen bonds (H-bonds) [which computes and analyzes hydrogen bonds which are determined based on cutoffs for the angle hydrogen-donor-acceptor and the distance donor-acceptor], using g_rms, g_rmsf and g_hbondgromacs analysis tools, respectively. H-bond was calculated when a donor-acceptor distance was less than 0.35 nm and a donor-hydrogen-acceptor angle was 30 degrees.
Investigation of interactions with PIC (protein interaction calculator)
The three dimensional structures of the SKC and the SKG132 were provided to Protein Interaction Calculation (PIC) server as an input to calculate various kinds of interactions such as H-bond, ionic interactions, interaction between hydrophobic residues, aromatic-aromatic interactions, aromatic-sulphur interactions and cation-pi interactions.24
Structure validation of the predicted models
To validate the quality of the modeled structures, Ramachandran map were generated using RAMPAGE Ramachandran plot assessment.16 Results of the Ramchandran plot for the best predicted model indicated that 86.8%, 9.7% and 3.5% of all the amino acid residues were in the favored, allowed and outlier regions, respectively; while very few residues could be found in disallowed region (Figure 2).

Figure 2 Ramachandran Plot validation for the best predicted model of SK structure. Ramchandran map were generated by Rampage validation program. Results indicated that 86.8%, 9.7% and 3.5% of all the amino acid residues were in the favored, allowed and outlier regions, respectively.
Molecular dynamics simulation
In order to determine key reasons of increased activity in SKG132, molecular dynamics simulation was carried out during 85 ns compared with SKC. RMSD of Cα atoms (Cα-RMSD) was analyzed with respect to minimized structure (Figure 3A). As illustrated in Figure 3A, the RMSD of both structures converge within the last 25 ns of simulation. Therefore, all of the analyses were performed through trajectories obtained from the last 25 ns of simulation. To examine the effect of mutations on the dynamic behavior of residues in SKG132, comparative analysis of the fluctuations (RMSF) of Cα atoms was calculated and shown in Figure 3B. As shown in Figure 3B, analysis of RMSF demonstrated remarkable changes of flexibility in three regions including 156-161, 183-187 and 301-372 in SKG132 compared to SKC. Based on Figure 3B, two regions including 156-161 and 183-187 showed increased flexibility in SKG132 compared to SKC, while the 301-372 region residues of SKγ in SKG132 exhibited an unexpected decrease in the flexibility compared to SKC. Structural examination of SKG132 confirmed that the 156-161 and 183-187 region residues are spatially located around the functionally important 250 loop which the more flexibility of these segments lead to better exposure of 250 loop against Pg substrate (Figure 4).
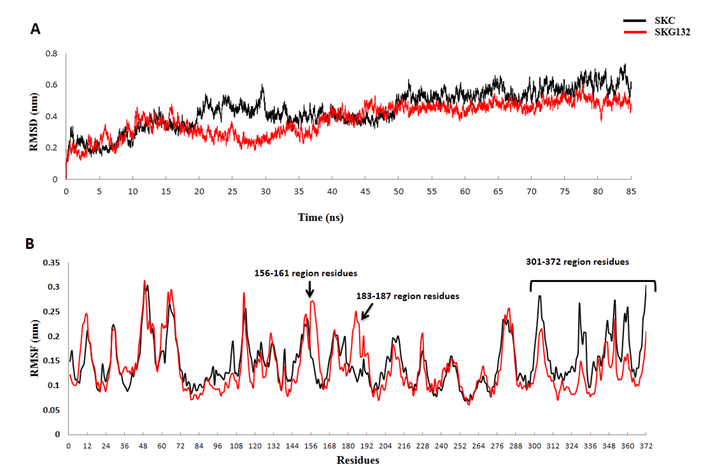
Figure 3 Evolution of the structural properties of the SKG132 and the SKC (black) during the MD simulations. Representation of the change in the root mean square deviation (RMSD) of C-alpha atoms (Cα-RMSD) of the SKG132 and SKC structures over the course of 85ns simulations with respect to the minimized structure. Cα-RMSD values of both structures converged within the last 25 ns of simulation, (B) The root mean square fluctuation (RMSF) of Cα atoms (Cα-RMSF) for both SKC132 and SKC were calculated from the last 25 ns structures of each simulation and values generally indicated significant changes of flexibility in three regions including 156-161, 183-187 and 301-372 in SKG132 compared to SKC. Specific regions in SK are indicated by arrows.

Figure 4 The cartoon representation of the enhanced flexible regions (156-161, 183-187) which are spatially located around 250-loop in SKG132. The 250 loop, 156-161 and 183-187 can be observed in ribbon and are colored in red, cyan and yellow, respectively. The structure is the average PDB structure of SKG132 which retrieved from the last 25 ns of MD simulations.
Hydrogen bond and PIC analyses
In order to clarify the relationship between flexibility and formation of H-bond, the number of H-bonds in three altered flexible regions (156-161, 183-187 and 301-372) were calculated during the last 25 ns of simulation. The average number of H-bonds in two increased flexible regions of 156-161 and 183-187 showed less number of H-bonds; 0.3 and 0.24 in SKG132, compared to the same regions in SKC with 0.45 and 1.8, respectively (Table 1).
Altered flexible |
Segment |
Average number of hydrogen |
Average number of |
||
SKC |
SKG132 |
SKC |
SKG132 |
||
156-161 |
6 |
0.45±0.05 |
0.3±0.03 |
0.08±0.001 |
0.05±0.002 |
183-187 |
5 |
1.82±0.07 |
0.24±0.06 |
0.36±0.07 |
0.05±0.001 |
301-372 |
72 |
50±4.81 |
53±4.67 |
0.69±0.08 |
0.74±0.06 |
Table 1 Comparison of average number of hydrogen bonds (H-bonds) and average number of H-bond per residue in three altered flexible regions in SKC and SKG132. H-bonds were calculated when a donor-acceptor distance was less than 0.35 nm and a donor-hydrogen- acceptor angle was 30 degrees. H-bonds were calculated by g_hbondgromacs built-in tools. All calculations corresponded to the last 25 ns of MD simulations.
The average number of H-bond per residue in 156-161 and 183-187 of SKC was 0.08 and 0.36, while it was 0.05 for both flexible regions in SKG132. Furthermore, the average number of H-bonds in 301-372 region of SKG132 was 53, while it was 50 in SKC. The average number of H-bond per residue for 301-372 region was 0.74 in SKG132 and 0.69 in SKC (Table 1).PIC server was used for computing intramolecular interactions of crucial residues for both SKC and SKG132 structures which achieved from the last 25 ns of simulation. Results of various interactions showed that the H-bond between Lys256 and Lys183 of (183-187) was disappeared in SKG132,while it was present in SKC with occupancy rate of 35% (Figure 5).24 Moreover, H-bond between residue pairs of Glu272-Lys256 and Glu272-Lys257 were also absent in SKG132, while they were existed in SKC with 94% and 100% occupancy, respectively (Figure 5).
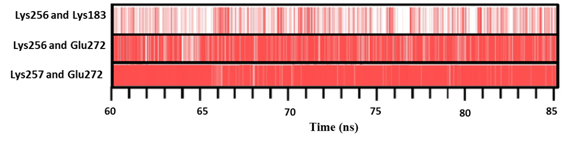
Figure 5 Hydrogen bonds (H-bonds) analysis in SKC and SKG132. Occupancy was calculated when a donor-acceptor distance was less than 0.35 nm and a donor-hydrogen-acceptor angle was 30 degrees. H-bond occupancy in residue pairs of Lys256-Lys183, Lys256-Glu272 andLys257-Glu272 in SKC was 35%, 94% and 100%, respectively, while it was absent in the SKG132. Calculations were corresponded to the last 25 ns of MD simulations.
In the present study, we used MD simulation of SKG132 and SKC to provide possible explanations of the role of residual substitutions, Ile33Phe, Arg45Gln, Asn228Lys and Phe287Ile, in high Pg activation of SKG132 compared to SKC. Our results showed increased flexibility of two regions including 156-161 and 183-187 and also decreased flexibility of 301-372 region in SKG132, which probably is the result of altered amino acid side chain interactions in substitution sites as demonstrated by PIC server and H-bond analysis.As shown in Figure 3B, RMSF analysis determined two enhanced flexible regions including 156-161 and 183-187 in SKβ of SKG132 relative to SKC. In prior studies, role of 250 loop and especially Lys256 and Lys257 at the tip of the loop are highlighted for substrate binding and recognition.4,8 Molecular docking study also indicated interaction between these lysine residues and K5 of Pg substrate.8 It should be noted that, two enhanced flexible regions of 156-161 and 183-189 are spatially located around of the 250 loop. Therefore, presence of these two flexible regions around 250 loop can improve exposure of 250 loop and particularly Lys256 and Lys257 for substrate binding and processing (Figure 4). Thus, better exposure of 250 loop and more efficient interactions with Pg substrate may contribute to high Pg activation of SKG132 relative to SKC.11 As shown in Figure 3B, 301-372 region residues of γ domain in SKG132 exhibited decreased flexibility. Declined flexibility of SKγ in SKG132 may result in formation of a stable activator complex. Previous studies also confirmed that, stable activator complex is prerequisite and essential for effective Pg substrate processing.9,25 Based on these studies, decreased flexibility of SKγ and consequently formation of a stable activator complex, may contribute with enhancement of SKG132 proteolytic activity which is in agreement with our experimental results.11As shown in Table 1, results of H-bond analysis in three altered flexible regions are in consistent with the RMSF value in related regions. Based on PIC results, H-bond interactions in residue pairs of Lys256-Lys183, Lys256-Glu272 and Lys257-Glu272 were disrupted in SKG132, while they were existed in SKC with high occupancy (Figure 5). Loss of H-bond between Lys256 and Lys183 could be one of the reasons for enhanced flexibility of 183-187 region residues. Two critical residues, Lys256 and Lys257, in 250 loop play an important role for Pg substrate processing.8 Therefore, disruption of H-bonds in three mentioned residue pairs (Lys256-Lys183, Lys256-Glu272 and Lys257-Glu272)led to generation of unbinding lys256 and Lys257, at the tip of 250 loop which can mediate better interactions with Pg substrate. In addition, due to the importance of 230-290 region for substrate interaction,4 generation of unbinding Glu272 in this region may also improve substrate recognition and processing in SKG132. Our results suggest that generation of unbinding critical residues; Lys256, Lys257 and Glu272 in SKG132 could potentially effect on Pg substrate interaction and consequently may result in high Pg activation of SKG132 which is in consistence with our experimental results.11 Therefore, we propose that flexibility of some critical regions and existence of some unbinding crucial residues might be vital for functional properties and alsoare contributed to the enhanced Pg potency of the SKG132 compared to that of the SKC.
Collectively, in the present study we introduced some aspects of high Pg activation in SKG132 compared to SKC using MD simulation. Our findings showed that the high Pg activation of SKG132 clearly mediated by increased flexibility of two regions including 156-161 and 183-187 and also a region with decreased flexibility in SKγ including 301-372. Presence of two increased flexible regions (156-161 and 183-187) around 250 loop and also generation of a stable activator complex due to decreased flexibility of a region in SKγ led to improved Pg processing. Moreover, generation of unbinding residues; Lys256, Lys257 and Glu272 which resulted from disruption of H-bond in residue pairs of (Lys256-Lys183, Lys256-Glu272 and Lys257-Glu272) may lead to better interactions with substrate and may play key role in increased activity of SKG132. Our results suggest that all four mutations in SKG132 (Ile33Phe, Arg45Gln, Asn228Lys, and Phe287Ile) might critically contribute as hot spots corresponding to structural changes toward increased Pg activation. This knowledge may be applied to make an SK molecule with designed properties for improving the clinical application of SK as a thrombolytic agent.
This work was supported by Pasteur Institute of Iran in fulfillment of the Ph.D. thesis of F. Kazemi.
Author declares that there are no conflicts of interest.

©2020 Kazemi, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.