MOJ
eISSN: 2374-6920


Research Article Volume 7 Issue 1
1Department of Applied Animal Science, Baba Bhimrao Ambedkar University, India
2Department of Zoology, University of Allahabad, India
Correspondence: Neeshma Jaiswal, Parasitology Laboratory, Department of Applied Animal Science, School of Bioscience and Biotechnology, Baba Bhimrao Ambedkar University, Lucknow-226 025, U.P., India
Received: June 28, 2017 | Published: February 19, 2018
Citation: Malviya S, Jaiswal N, Malhotra SK. Molecular diagnostic procedures using RT-PCR to alleviate taxonomic impediments of parasite species segregation. MOJ Proteomics Bioinform. 2018;7(1):53-56. DOI: 10.15406/mojpb.2018.07.00213
The taxonomic impediments in taxa differentiation have been overcome by the application of advanced techniques using real time PCR methods. PCR product detection and identification of helminth species is performed with a variety of fluorescence chemistries. The measurement of melting temperatures (Tms) of amplicons, after completion of PCR, facilitated precise diagnosis of species of particular helminth. The technique is reliable as the Tms of the specific amplicons are sufficiently different and constant to illustrate differentiation. The phylogeny of Contrcaecum sp. from a marine fish, Sillago sihama is established with report of Tms, 87.5°C, using melting curve analysis real-time PCR in the present investigation, with simultaneous segregation of Anisakis sp. as a separate taxon from the coexisting species, Johnius dussumieri whose genotypes have been reported at the GenBank.
Keywords: real time PCR, molecular diagnostic procedures, melt point temperature, contrcaecum bancrofti, anisakis sp
The development of reliable tools for surveillance, diagnosis and detection of such diseases at a quicker pace has indeed been challenging in context of the emerging infectious diseases, particularly in the tropical regions of the world. The primary requirement of an effective tool of such diagnosis is the skill of a tool to detect absence of neglected tropical diseases. Meltpoint DNA-based diagnostics are being developed for a number of diseases and although qRT-PCR is the most common detection method at present, there is extensive interest in improving and diversifying detection technologies, which may provide more field-friendly tools. The technicalities of biotechnological tools, appropriateness of technological application, and rapid adaptability, as according to the alterations and adjustments suited to the progress of the control programs through different phases, from the peak prevalence of infections to the definitive diagnostics to the infections that have disappeared. The preference for an easier to use tool that had a cost-effective efficiency was propagated1,2 even if it bore a moderate sensitivity. This was particularly because rapidity of mapping to mark high priority regions, where infection prevalence was high and frequency of screening at a brisk pace was the requirement. The sensitivities and specificities of comparable diagnostic tests were considerably improved after introduction of PCR oriented estimations and analysis before which the methods incorporating meta-analyses of diagnostic test proficiencies were invariably relied upon, examples of which are commonly available in cases for the assessment of Chagas disease, leishmaniasis and malaria,3–5 as well as Soil Transmitted Health (STH) diagnostic technology.6–9 Some of the most common techniques for detection and diagnosis of Ascaris lumbricoides, Trichuris trichiura and Ancylostoma duodenale involving Kato-Katz,10 direct microscopy,11 formol-ether concentration (FEC),12 McMaster,13 FLOTAC14 and Mini-FLOTAC15 methodology. In recent years, the sensitivity of different diagnostic tests16,17 was assessed by the application of Bayesian latent class model.18
The marine fish Sillago sihama from Arabian Sea at the Central west coast of Goa, yielded eightynine specimens of larval Contrcaecum in the intestine of fish. Simultaneously the mature worms of Anisakis sp. were recovered from a sciaenid, Johnius dussumieri, during February through November, 2014. The method of genomic DNA extraction and determination of the Melt point temperature of a sciaenid, J. dussumieri, respectively, during February through November, 2014 were as described already in an earlier publication19 using RT-PCR technology. The rDNA region comprising the ITS1 and ITS2 sequences was amplified with primers, SS1, 5’-GTTTCCGTAGGTGAACCTGCG-3’; and NC13R, 5’-GCTGCGTTCTTCATCGAT-3’ for ITS1 gene. Total DNA was extracted from these worms, using DNeasy Tissue Kit (Qiagen) in a final buffer volume of 50um. A 950 base pair (bp) fragment of the Inter Transcribed Spacer –1 gene from the extracted DNA was amplified using primers19 and a Thermal Cycler IQ5 Real Time PCR Detection System Biorad Laboratories Inc., Hercules, CA, USA.20 2-way ANOVA was employed for two-factor differential diagnostic analysis.
The protozoan infections of zoonotic significance, that were characterized by asymptotic patterns, were detected in human beings.21,22 The causative agents of helminth infections of marine and freshwater fish from Indian aquatic ecosystems were identified by illustrating their strikingly differentiable melt point peak temperatures.19 The specific melt point temperature curves i.e. derivative melt curves (Figure 1a) and aligned melt curves (Figure 1b) were worked out in the current investigation to illustrate distinguishing feature of larva of Contracaecum bancrofti (Figure 1a). The unique melt point temperature peak of 87.0°C±0.01°C that was available by the application of real-time PCR with SYBR Green1 added identifiable characteristics by using advanced technique. The findings of experimentation to denote peaks of melting point curves of ITS2 gene to distinctly establish the separate identity of adult specimens of A. simplex in derivative melt curve (Figure 2, 79.59°C±0.02°C), and in aligned melt curve from J. dussumieri are shown in Figures 2 & 3. Running a 2-way ANOVA analysis, we found that the melt point temperatures of Contracaecum were significantly different from that of Anisakis, (P < 0.05), and the power of the test was 80%.
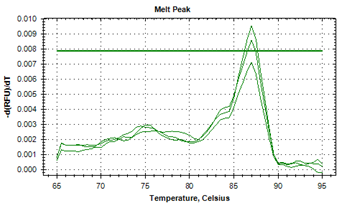
Figure 1a Profile of the melt peak curves (derivative melt curves) showing denaturation profile of ITS1 amplicons in three replicates, for larval Contracaecum bancrofti (KF990496) recoveredfrom Sillago sihama.
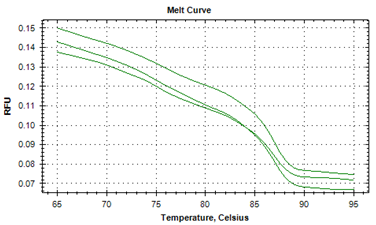
Figure 1b Profile of the melt peak curves (aligned melt curves) showing denaturation profile of ITS1 amplicons in three replicates, for larval Contracaecum bancrofti (KF990496) recoveredfrom Sillago sihama.
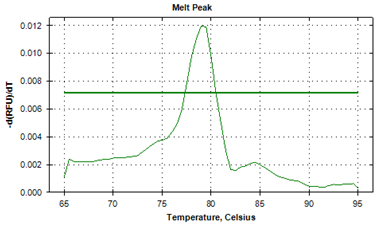
Figure 2 Profile of the melt peak curves (derivative melt curves) showing denaturation profile of ITS1 amplicons in three replicates, for larval Contracaecum bancrofti (KF672839) recoveredfrom Sillago sihama.
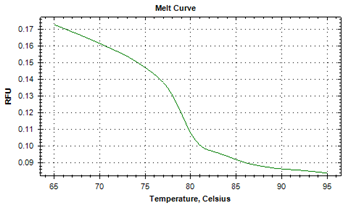
Figure 3 Profile of the melt peak curves (aligned melt curves) showing denaturation profile of ITS1 amplicons in three replicates, for adult Anisakis sp. (KF672839) recoveredfrom Johnius dussumieri.
Conclusively, the anisakid, A. simplex, had lower Tm values. The melting characteristics of ITS1 amplicons from all species were assessed by plotting two different curves (Figures 1a, 1b, 2 & 3). In the present study, the normalized fluorescence curves i.e. aligned melt curve (Figures 1b & 3) and derivative melt curve (Figure 1a & 2) produced uniquely different plots that were easily distinguishable for each species. It would mean that although the melting profiles of different species (Tm) were very close to each other, they could clearly be discerned by the plotting of normalized melting curves (Figures 1a , 1b, 2 & 3). It has been recently asserted23 that for species having almost similar melting curves and temperature-shifted fluorescence difference, a sharp decrease in fluorescence was detected in denatured DNA that was consistent for such species, with its respective melting point. The binding of intercalating dyes to any double stranded DNA is its drawback and henceforth melting point curve is a point for analysis, as non specific DNA is denatured at this point and specific DNA products remain intact. This associates the melting point to the composition and sequence of the nucleotides and characterize sequence variation within the samples. The specific diagnosis made by the melting point curve is of importance in elucidating the genetic diversity, and will assist in study of differences in the biology, ecology and in the transmission of the parasites.24
The specific melt point temperature curves i.e. derivative melt curves (Figure 1a) and aligned melt curves (Figure 1b) were worked out in the current investigation to illustrate distinguishing feature of larva of Contracaecum Bancroft (Figure 1). The unique melt point temperature peak of 87.0°C that was available by the application of real-time PCR with SYBR Green1 added identifiable characteristics by using advanced technique. The cracking out of phylogeny of these worms provided evidence of alignment of ITS1 sequence of larval Contracaecum bancrofti (KF990496) with the sequence (EU839573) submitted with the GenBank by the earlier authors, of adult C. bancrofti (showing bootstrap value 100) (Figure 2). The monophyletic association between the sequences of Contracaecum spp. was evident. This reliably emphasized the larval forms of worms investigated in this study, to be the larvae of C. bancrofti recovered from Indian marine fish, S. sihama. The investigation thus very well established that the molecular diagnostic procedures incorporating RT-PCR are a dependable method to achieve species level diagnostics, that could be critical to deal with parasitic helminthes of pathogenic significance.
The ITS1 sequence of the other nematode, Anisakis sp. (KF672839) whose sequence was analysed in this study, did not align with any other nucleic acid sequence of other known Anisakis spp. (Figure 2) submitted at the GenBank uptill now, and instead stood apart entirely to suggest it to be a separate taxonomic entity among anisakids that infected fish at the Arabian Sea at Central west coast of India at Goa. Therefore, a polyphyletic association, entirely separate from the two groups- 1. of Contracaecum spp. and 2. of Anisakis spp. was concluded.
The implications of phylogenetic allegiance could well be linked to the issue of melting point of amplicon being a factor of nucleic acid sequence length that could be sensitive to highlight the emerging specificity of an individual helminth, which is critical to taxa differentiation. This evidently didn’t allow alignment of Anisakis sp. (KF672839) with any of the available sequences with GenBank (Figure 2). It is strongly asserted that these advanced methods could be helpful to remove impediments to ward off ambiguity in species level segregation of taxonomic entities.
A number of species of intestinal helminths are the cause of certain most common infections occurring in human beings in various states of India. Necator americanus and Ancylostoma duodenale add to the list of hookworms as well. In addition to these, canine as well as feline hookworms, A. ceylanicum, A. caninum and A. braziliense also have occurrence worldwide.25–27 The noticeable debilitating effect on the socio-economic conditions of humans occurred due to the chronic loss of blood in the intestinal tissues, that could trigger iron deficiency, anaemia and hypoalbuminemia28,29 that, in turn, impaired physical, intellectual and cognitive development of adolescents and enhanced premature deaths during pregnancy.30–34 The formulation of effective therapeutic measures to control hookworm and other helminth infections was dependent on accurate identification of worms and their molecular characterization of these worms. The recent focus on methods of diagnostic application for nematodes encompassed specific assessment of the specificity of genetic markers such as first (ITS1) and second (ITS2) internal transcribed spacer of nuclear ribosomal DNA (rDNA), using advanced molecular techniques.35,36 The application of conventional and semi-nested PCR and single-strand conformation polymorphism (SSCP),37 mutation scanning38 and PCR-restriction fragment length polymorphism (RFLP)39 were illustrated in earlier years.36 Therefore, to avoid time consuming, and costly electrophoretic analysis, the advanced diagnostic techniques, including the rapid, high-throughput diagnosis and genetic analysis of pathogens as well as data handling and analysis are being employed in current times. Of late the emphasis on High-resolution melting (HRM) in clinical investigations40–44 for rapid screening and segregation of closely related species has frequently been laid on the studies45 on Brugia malayi and B. pahangi, Fascioloides magna, Leishmania spp.,45 Cryptosporidium spp.,46 Plasmodium falciparum,47 Dientamoeba fragilis,48 Naegleria spp.49 and Giardia spp.50
Hence RT-PCR is a unique method enabling relative or absolute quantification of nucleic acids prior to which the quantification of cyclic events was essential to be through beyond the threshold during commencement of the amplification cycle.
SJM is thankful to the Principal, Govt. Postgraduate College, Naini (Allahabad) for facilities to carry out the current research work. The authors are grateful to Professor Sandeep K. Malhotra Former Head, Department of Zoology, University of Allahabad for RT-PCR technology facilities in the Department.
The authors declare that there is no conflict of interest regarding the publication of this article.

©2018 Malviya, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.