MOJ
eISSN: 2374-6920


Review Article Volume 2 Issue 4
1Department of Experimental Therapeutics, University of Texas M.D. Anderson Cancer Center, USA
2Department of Head and Neck Surgery, University of Texas M.D. Anderson Cancer Center, USA
3Department of Surgical Oncology, University of Texas M.D. Anderson Cancer Center, USA
4Windsor University School of Medicine, St. Kitts, West Indies
5Department of Medicine, Section of Emergency Medicine, Baylor College of Medicine, USA
Correspondence: Arumugam Jayakumar, Department of Experimental Therapeutics, The University of Texas, USA
Received: July 22, 2015 | Published: August 20, 2015
Citation: Jayakumar A, Frederick M, Shellenberger T, et al. Molecular and biochemical properties of lympho-epithelial kazal-type-inhibitor (LEKTI). MOJ Proteomics Bioinform. 2015;2(4):121-127. DOI: 10.15406/mojpb.2015.02.00053
Here, we review the literature on the molecular and biochemical properties of Lympho-Epithelial Kazal-Type-Inhibitor (LEKTI). Serine Protease Inhibitor Kazal-type 5 (SPINK5) gene encodes three different LEKTI isoforms. These isoforms are organized into a typical 15, longer than 15 and shorter than 15 inhibitory domains consisting of 1064, 1094 and 916 residues respectively. LEKTI isoforms synthesized as pro-LEKTI proteins are processed intracellular and secreted as bioactive LEKTI fragments into blood. LEKTI potently inhibits activity of plasmin, subtilisin A, cathepsin G, elastase, trypsin, kallikrein (KLK) 5, KLK6, KLK7, KLK13, and KLK14 to varied extents. Mutations in SPINK5 gene resulting in decreased LEKTI function and increased KLK activity causes Netherton syndrome (NS). Low SPINK5 expression/LEKTI function is also associated with Head and Neck Squamous Cell carcinoma (HNSCC), chronic rhinosinusitis and asthma. Thus restoring SPINK5 expression/LEKTI function in NS, HNSCC and asthma holds therapeutic promise.
Keywords: SPINK5, LEKTI, LEKTI, domains, proteinases, NS, HNSCC, furi
LEKTI, lympho-epithelial kazal-type-inhibitor; SPINK5, serine protease inhibitor kazal-type 5; KLK, kallikrein; NS, netherton syndrome; HNSCC, head and neck squamous cell carcinoma
SPINK5 gene was initially cloned following sequence identification of two polypeptides, HF6478 and HF7556, isolated from human blood filtrates.1,2 It was also independently cloned as the genetic locus responsible for Netherton syndrome.3 Owing to the presence of Kazal-type domains in the translated protein and its expression pattern in different organs, the gene encoding these polypeptides was named Lympho-Epithelial Kazal-Type-related Inhibitor (LEKTI).4 It was shown for several other endogenous proteinase inhibitors like tissue inhibitors of matrix metalloproteases (TIMPs), maspin, elafin, hespin, headpin, SERPINs, and SPI that they regulate the proteolytic signaling involved in homeostatic and disease processes.5–12 Since the identification and cloning of SPINK5 substantial work has been done describing the targets and biologic roles of its gene product LEKTI. The purpose of this chapter is to review the current literature on the molecular and biochemical properties of LEKTI.
Identification and molecular cloning of SPINK5 cDNA
Since the original report of the cloning of SPINK5 gene,1 several groups including us have reported the identification and cloning of a total of three isoforms of SPINK5 gene encoding 3 LEKTI isoforms.13–18 These three isoforms are: a typical LEKTI containing 15 domain (1064aa), a shorter LEKTI containing only first 13 domains (916aa) and a longer LEKTI with a 30-amino-acid insertion between domains 13 and 14 (1094aa) (Figure 1). We have demonstrated that SPINK5 gene is one of the nine genes down-regulated in Head and Neck Squamous Cell Carcinoma (HNSCC). Subsequently, we have cloned LEKTI cDNA encoding LEKTI protein consisting of1064 residues from normal oral mucosa utilizing LEKTI specific primers and RNA isolated from normal oral mucosa.19 We have separately amplified the 5' (1.8-kilobase) and 3' (1.3-kilo base) halves of the LEKTI cDNA and then sub cloned these two fragments into pCRII-TOPO vector. Sequencing the entire LEKTI cDNA found out six silent single base exchanges in the open reading frame compared to reported LEKTI sequences (GenBank/EMBL accession no. AJ228139 and AF086524). On the basis of the open reading frame we have predicted that the deduced amino acid sequence of 1064 residues with a Mr of 121,234. A large body of literature has demonstrated that loss-of-function mutations in SPINK5 gene cause Netherton syndrome.3,20–24 A recent report has discovered a novel putative long non-coding RNA (lncRNA), which is antisense to SPINK5 gene,25 implicating a translational regulation of SPINK5 gene expression at least in some tissues.
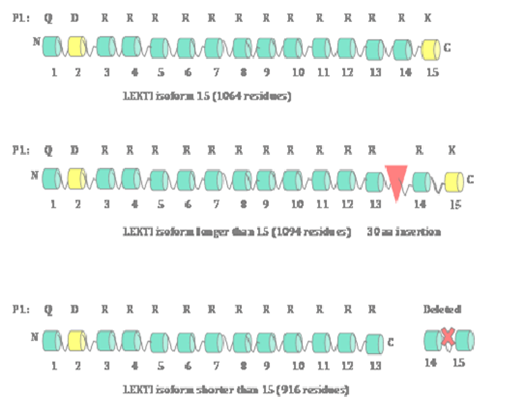
LEKTI organization
On the basis of the furin cleavage sites found within the LEKTI polypeptide containing1064 amino acids, there are 15 potential inhibitory domains (Figure 1). There is a secretory signal peptide sequence containing 22 amino acids at the N-terminal of the full-length poly-peptide.26 It was also established that LEKTI domains 2 and 15 resemble typical Kazal-type serine proteinase inhibitors with 3 disulfide bridges whereas domains 1, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13 and 14 resemble non-Kazal-type inhibitors with only two disulfide bridges.27–30
Expression and purification of human LEKTI in insect cells and E.coli
We engineered a recombinant baculovirus expression vector encoding the entire native human LEKTI protein (including the putative signal peptide) fused in frame with a C-terminal six-histidine tag (Figure 2). We could not detect a secreted form of LEKTI when expressed in Sf9 cells consistent with previous reports that the N-terminal leader sequences of proteins of higher eukaryotes often yield only small quantities of secreted proteins in yeast and insect cells. We did not attempt to improve the secretion efficiency in Sf9 by replacing or fusing the LEKTI native signal sequence with an insect cell signal sequence (e.g. melletin). Instead, we selectively purified the LEKTI precursor from cell lysates. Following infection of Sf9 with recombinant baculovirus, abundant rLEKTI was detected in cell lysates by immunoblotting with penta His mAb. We processed the cell lysates by TALON metal affinity and gel-filtration chromatography as previously described.12 Upon purification, a sole band of »120kDa was visualized by Coomassie brilliant blue R-250staining in the pooled imidazole eluates (Figure 2). N-terminal sequencing of the protein band produced no amino acid sequence, which suggested that the N-terminal residue may have been blocked in vivo or modified during our sample preparation. Internal amino acid sequencing of this protein band confirmed it to be bona fide LEKTI. Approximately 0.7mg of pure LEKTI was obtained from the Sf9 cell pellet of a one-liter culture. These results suggested that C-terminal six-histidine tagged rLEKTI could be efficiently expressed and selectively purified from insect cells. In order to evaluate whether rLEKTI contained disulfide bonds and to determine whether protein aggregates were present in our preparation, rLEKTI was analyzed by SDS-PAGE under both reducing and non-reducing conditions. The rLEKTI separated by SDS-PAGE under non reducing conditions clearly migrated faster than under reducing conditions providing evidence of disulfide bonds present in the recombinant protein. This clearly indicates that almost all of the LEKTI expressed in insect cell contained disulfide bonds and this Sf9 produced rLEKTI could be used to screen for inhibitory activity.
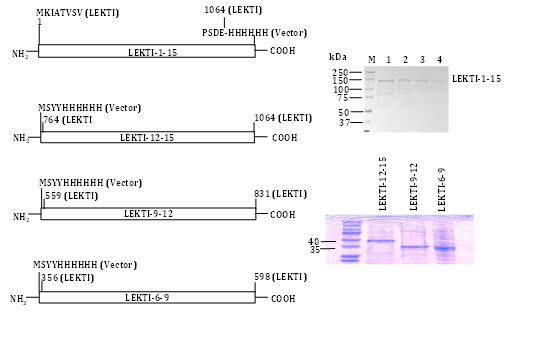
Having successfully purified pro-LEKTI in insect cells we also engineered LEKTI multi domains 1-6, 6-9, 9-12, and 12-15 expression composite bacmids, expressed, and purified these proteins in Sf9 cells.31 The expression constructs and their corresponding purified LEKTI multi domains except for LEKTI domains 1-6 are shown in Figure 2. Other research groups have reported the expression and purification of LEKTI single domains 6 and 15 using a bacterial expression system.29,32 Testing a selected number of different serine proteinases, the authors have shown that both native and recombinant LD-6 exhibit a significant but temporary inhibitory activity on trypsin.
Genomic organization and expression status of LEKTI in normal and Comèl-Netherton syndrome patients
Magert et al.,32 have shown for the first time that SPINK5 encoding LEKTI was localized on human chromosome 5q31-32.1 We and others have demonstrated that LEKTI/SPINK5 gene is expressed in the normal oral mucosa and normal pituitary glands,16,33 thymus, vaginal epithelium, Bartholin's glands, , tonsils, and the parathyroid glands.1 For the first time, Hovanaian and his group reported different mutations in SPINK5 in families with Netherton syndrome (NS, MIM256500).3,13,34 Subsequently the same group described the intron-exon organization of SPINK5 gene and characterized its mutations in patients from 21 families of different geographic origin, using denaturing high performance liquid chromatography and direct sequencing.15,35 These results have collectively shown that most of these mutations predict premature termination codons resulting in diminished LEKTI expression. Subsequent work by other investigators confirmed and extended that SPINK5 mutations do occur in Japanese, Taiwanese, Chinese, Korean, Turkish and Israeli populations.36–42 In our own studies we have shown that although the absence of LEKTI proteins in NS patients lead to over desquamation, there is a compensatory upregulation of DSG3/DSC3 in NS patients permitting survival of these patients in the face of unregulated hKLK activity.22,43
Genome-wide and microarray analyses of SPINK5/LEKTI in HNSCC, asthma and allergies
A Genome-wide transcriptomic profiles obtained for 53 primary oral tongue squamous cell carcinoma (OTSC Cs) and 22 matching normal tissues identified SPINK5 as one of the genes down-regulated in OTSCCs.44 Their Gene Ontology analysis discovered an increases phosphate transport, collagen catabolism, NF-kB signaling cascade, extracellular matrix organization and biogenesis, chemotaxis, as well as suppressions of superoxide release, and hydrogen peroxide metabolism, in OTSCCs. In a recent study Shah et al. generated a transcriptome map of BMSCC cancer, performed RNA-seq analysis, and assessed the role of alternative splicing in BMSCC. They detected a total of 11 novel splice junctions derived mostly from alternate 5' splice site and also found novel putative long non-coding RNA (lncRNA), which is antisense to SPINK5 gene.45 A recent report correlated the clinicopathological parameters of 83Non-HPV OSCC patients with the protein expression for KLK5, KLK7 and LEKTI in these tumors and demonstrated that concurrent loss of KLK5 and KLK7 associates with a poor clinical outcome in OSCC and could therefore serve as prognostic marker in this disease.46 A recent study also evaluated whether single-nucleotide polymorphisms or haplotypes at 5q31-33 conferred risk for asthma in Mexican-Mestizo pediatric patients.47 They utilized TaqMan Allelic Discrimination Assay and genotyped 20 single-nucleotide polymorphisms within , RAD50, IL13, IL4, CD14, SPINK5, HTR4, ADRB2 and IL12B. Their results revealed that three SPINK5 haplotypes (GGCT: p=6 × 10(-6); aaTC: p=0.0001; AGTT: p=0.0001) were associated with asthma. We recently compared the expression profile of about 18,000 genes and identified 186 genes which are differentially regulated between LEKTI stable clones in comparison with the vector transfected OSC19 parental cells. Among them, mMP-14,mMP-8, KLK5, and ADAM8 are down regulated andmMP-3, LEKTI, DSC2 and DSC3 are up-regulated in LEKTI clones.48
Mass spectrometry analyses of recombinant LEKTI
In Lauber et al.,27 reported for the first time the cloning, over expression, purification, and mass spectrometry analysis of recombinant LEKTI domain (HF6478) in Escherichia coli Origami (DE3) strain which carries a trxB(-)/gor522(-) double mutation.27 They have demonstrated that the molecular weight of this protein as obtained by electro spray mass spectrometry (6477.69) was in very close agreement with 7kDa apparent molecular weight seen in SDS-PAGE. In the same year, Ahmed et al have purified proteins/peptides from concentrated growth medium of epidermal keratinocytes and sequenced by Edman degradation in a gas-phase sequencing system. Their analyses have shown that the 30 and 40kDa proteins sequenced up to 18 and 21 residues respectively are in alignment with human LEKTI and DAN proteins.49 Subsequently, in 2003, we have purified full length rLEKTI and performed the N-terminal and internal amino acid sequence analyses.19 Our results shown for the first time that the N-terminal sequencing of the protein band produced no amino acid sequence suggesting that the N-terminal residue may have been blocked in vivo or modified during our sample preparation. However, internal amino acid sequencing of this protein band confirmed it to be bona fide LEKTI. In addition, we performed digestion of rLEKTI with human recombinant furin and the resulting cleavage fragments were resolved by SDS-PAGE and MALDI-MS.19 SDS-PAGE analysis showed that furin cleaved LEKTI into six discrete polypeptides ranging in molecular mass from 10kDa to 40kDa. However, the MALDI-MS analysis for polypeptides in 6000 to 9000 Da ranges detected the presence of 13 major peptide peaks.
Recombinant LEKTI inhibit a broad spectrum of proteinases
Using purified pro-LEKTI and different LEKTI multi domains, we and others tested their inhibitory activities against a panel of proteinases.19,29,31,50-54 In collaboration with Dr. Schechter, we studied the effects of LEKTI domains 6-9 and 9-12 on the in vitro activity and binding of human recombinant KLK5 and KLK7.50 We demonstrated for the first time that LEKTI domains 6-9 strongly inhibited the activity of both KLK5 and KLK7 with a Ki of 5nM and 11nM respectively. In contrast, LEKTI domains 9-12 inhibited KLK5 only with a Ki of 3nM but not KLK7. We have also shown that this inhibition is almost the same irrespective of the pH 8.0 and pH 5.0 at which both the binding and kinetic studies were performed. Our results further suggested that the LEKTI bioactive peptides are most likely very active in stratum corneum where both LEKTI and proteases are bathed at an acidic pH. In our later studies and in collaboration with Drs. Deraison and Diamandis,52,55 we have determined the effects of four LEKTI domains including two new LEKTI domains 1-6 and LEKTI domains 12-15 on the in vitro activity of a panel of human KLKs including KLK1, KLK5, KLK6, KLK13 and KLK14 (Table 1).
rLEKTI Species |
Protease |
Inhibition (%) |
Ki (nM) |
Mechanism (s) |
References |
12-Jan |
Plasmin |
97 |
27 |
Non-Competitive (NC) |
|
1-12,6-9 |
Subtilisin A |
80 |
49,350 |
NC |
|
12-Jan |
Cathepsin G |
85 |
67 |
NC |
|
12-Jan |
HNE |
90 |
300 |
NC |
|
1-12,6-9 |
trypsin |
90 |
800,200 |
NC |
|
1-6,6-9,9-12,12-15 |
KLK1 |
0 |
- |
- |
|
1-6,6-9,9-12,12-15 |
KLK5 |
90 |
2,5,3,22 |
Mixed |
|
1-6,6-9,9-12, |
KLK6 |
90 |
13,48,195 |
Mixed, NC,NC |
|
6-9 |
KLK7 |
90 |
11 |
Not Determined |
|
1-6,6-9,9-12, |
KLK13 |
90 |
24,222,409 |
Mixed, NC,NC |
|
1-6,6-9,9-12, |
KLK14 |
90 |
0.22,3,10 |
Mixed |
|
1-12 |
chymotrypsin |
0 |
- |
- |
|
1-12 |
Papain |
0 |
- |
- |
|
9-Jun |
Chymase |
0 |
- |
- |
|
1-12 |
Cathepsin K |
0 |
- |
- |
|
12-Jan |
Cathepsin L |
0 |
- |
- |
|
1-12 |
Cathepsin s |
0 |
- |
- |
Table 1 Sensitivity of various proteases to inhibition by rLEKTI
Our collective results have shown that LEKTI1-12 and LEKTI multidomains had a strong inhibitory effect on trypsin, plasmin, KLK5, KLK6, KLK13, KLK14, cathepsin G, HNE, and subtilisin A. They had no inhibitory effect on KLK1, chymase, chymotrypsin and cysteine proteinases papain or cathepsins K, L, or S (Table 1). To understand whether LEKTI behaves as a slow - or fast-binding inhibitor, we measured the time course of various proteinase activities in the presence of different concentrations of rLEKTI. We have observed that the product formation over the 60 min assay period in the absence and presence of inhibitor was linear with respect to time.19,31,49,54 The linear shapes of these inhibition curves indicate that rLEKTI is not a time-dependent inhibitor, suggesting that LEKTI binds rapidly to these proteinases and inactivates them. To classify the type of inhibition, the kinetic constants (Km and Vmax) of plasmin, trypsin, subtilisin A, cathepsin G, HNE, KLK5, KLK6, KLK13 and KLK14 were determined for their respective chromogenic or flurogenic peptides in the presence of increasing concentrations of different rLEKTI species.19,31,55 The Eadie-Hofstee plots (V/[S] versus V) show that apparent Km remain same or increased while apparent Vmax decreased, as [LEKTI] increased, indicating that the inhibition was either non-competitive or mixed. On the basis of the Ki values, rLEKTI1-12 is a potent non-competitive inhibitor of plasmin (27nM), subtilisin A (49nM), and cathepsin G (67nM), and rLEKTI1-12 is only a moderate inhibitor of HNE (300nM), however, and a weak inhibitor of trypsin (800nM).
Our comprehensive analyses have also established that none of the four LEKTI species we have tested inhibited KLK1 activity. We observed that LEKTI domains 1-6, 6-9, 9-12, and 12-15 is a potent mixed type inhibitor of KLK5 with a Ki of 2nM, 5nM, 3nM, and 22nM. We also demonstrated that LEKTI domains 12-15 did not inhibit the activity of KLK6, KLK7, KLK13, and KLK14. LEKTI domains 1-6 is a potent mixed and LEKTI domains 6-9 and 9-12 are a potent non-competitive inhibitor of KLK6 with a Ki of 13nM and 48 and 195nM respectively. Similarly, LEKTI domains 1-6 is a potent mixed and LEKTI domains 6-9 and 9-12 are a potent non-competitive inhibitor of KLK13 with a Ki of 24nM and 222 and 409nM respectively. Finally, all three LEKTI domains 1-6, 6-9, and 9-12 showed a mixed type of inhibition for KLK14 with a Ki of 0.22nM, 3nM, and 10nM respectively.
Among the KLK family, KLK5 was shown to degrade desmoglein-1 (DSG1) in skin.18 DSG1 stability is very important for skin function. Previously, it was shown that production of LEKTI domains 6-9 was compromised due to a mutation of E420K in atopic dermatitis (AD) populations56,57 implicating that variants in SPINK5 may be associated with asthma and allergy. Consistent with these observations, a recent study demonstrated that double knockdowns of SPINK5 and KLK5 in normal human epidermal keratinocytes by small interfering RNA (siRNA) lead to an increased expression of desmocollin 1 (DSC1), desmoglein 1 (DSG1) and (pro)filaggrin suggesting that inhibition of serine proteases KLK5 and KLK7 could be therapeutically beneficial in NS.58 Collectively, our results have established a new mechanism of skin homeostasis via regulation of skin KLKs activity by LEKTI.
Disulfide is required for LEKTI inhibitory activity
We have examined the influence of dithiothreitol (DTT), a reducing agent, on the rLEKTI 1-12 inhibitory activity of plasmin and subtilisin A. We have demonstrated that in the absence of a reducing agent, rLEKTI1-12 inhibitory activity was constant at pH 7.8 and room temperature for 1h.14 In contrast, rLEKTI1-12 inhibitory activity of plasmin and subtilisin A was readily inactivated by DTT (20mM) during this time. In control experiments, the addition of DTT alone had no effect on plasmin and subtilisin an activity. Therefore, the reduction in rLEKTI inhibitory activity was not due to an artificial increase in proteinase activity by the presence of DTT.
LEKTI processing and secretion
Although endogenous LEKTI full length proteins were synthesized as one of the121kDa pro-LEKTI containing intact 12 domains, Ahmed and Magert reported the isolation of LEKTI individual domains 1, 5, and 6 and LEKTI multi domains 8-12 from human blood filtrate and conditioned medium of epidermal human keratinocytes respectively.4,49 These observations suggested that full length LEKTI is subject to both extracellular and intracellular proteolysis by some unknown proteases. To understand more about this phenomenon, we analyzed the stability and subcellular distribution of several LEKTI deletion mutants utilizing HEK 293T cells as expression host. Our results have established that furin is required for the LEKTI intracellular processing in vivo resulting in three cleavage products of 37-, 40-, and 60kDa LEKTI.59 Furthermore, on the basis of these findings, we suggested two potential furin cleavage sites, one around LEKTI residue 352 and the second one around LEKTI residue 678. Consistent with our results Fortugno et al. identified three LEKTI fragments and showed a positive correlation between the quantity of LEKTI polypeptides and activity of KLK5 in the epidermis.60 In addition to furin as a LEKTI processing enzyme, we recently identified meprins also involved in the processing of intracellular LEKTI.61 It was also recently reported that mesotrypsin enzyme localized in the granular cells could degrade LEKTI and indirectly activate skin KLKs.62
Intracellular furin cleaves cellular LEKTI
To further elucidate the role of furin, we have recently transfected human colon carcinoma LoVo cells deficient in furin with pro-LEKTI alone or pro-LEKTI plus furin and determined the expression and processing of LEKTI in cell lysates and conditioned medium. Our results showed that in the absence of furin expression no LEKTI processing was observed in LoVo cells. However, LEKTI is readily processed upon furin expression confirming and extending our previous results that processing of recombinant LEKTI is furin-dependent (Figure 3).
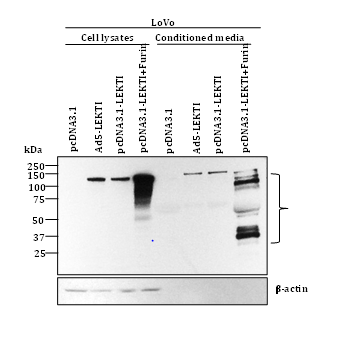
Funding
Supported in part by the NIH-NCI P50 CA097007, NIH R01 DE013954, NIH P30 CA016672, Alando J. Ballantyne Distinguished Chair in Head and Neck Surgery award, Michael A. O’Bannon Endowment for Cancer Research, NIH INRS Award T32 CA060374, andaaO-HNSF Percy Memorial Grant to G.C.
A.J and G.C provided intellectual input into the design and presentation of the study. A.J wrote the manuscript. MF, KB, TS, YH, KJ, and YK carried out most of the experiments. V.R. and K. J reviewed LEKTI literature and organized the data.
The author declares no conflict of interest.

©2015 Jayakumar, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.