MOJ
eISSN: 2374-6920


Research Article Volume 1 Issue 3
1Department of Head and Neck Surgery, University of Texas MD Anderson Cancer Center, USA
2Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, USA
3Department of Experimental Radiation Oncology, University of Texas MD Anderson Cancer Center, USA
Correspondence: Arumugam Jayakumar, Department of Head and Neck Surgery, University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA
Received: July 01, 2014 | Published: July 8, 2014
Citation: Jayakumar A, Chattopadhyay C, Wu HK, et al. LEKTI, a physiological inhibitor of multiple serine proteinases, blocks migration and invasion of head and neck squamous cell carcinoma (HNSCC) cells. MOJ Proteomics Bioinform. 2014;1(3):57-71. DOI: 10.15406/mojpb.2014.01.00015
Serine Protease Inhibitor Kazal-type 5 (SPINK5) gene encodes 3 different Lympho-Epithelial Kazal-Type-Inhibitor (LEKTI) isoforms which are organized into longer than 15, 15, and 13 inhibitory domains. We identified LEKTI by its constitutive expression in normal oral mucosa and lost or down regulated expression in matched tumor specimens of patients with head and neck squamous cell carcinoma (HNSCC). Previously, we showed that recombinant full-length LEKTI and rLEKTI fragments inhibit the activity of plasmin, subtilisin A, cathepsin G, neutrophil elastase, trypsin, caspase 14, and kallikreins (KLK) 5, 6, 7, 13, and 14 to varied extents. Here, we show that LEKTI protein is absent in HNSCC OSC19, Tu138, Tu177, and UMSCC1 lines. We then determined the consequences of LEKTI re-expression on migration and invasion, adhesion and gene expression profile of HNSCCOSC19 and UMSCC1 lines. We demonstrate that LEKTI expressing OSC19 and UMSCC1 clones show markedly reduced migration and invasion. Moreover, LEKTI expressing OSC19 clones show striking morphological changes and enhanced adhesionon type I, III, IV, and V collagens, fibronectin, and laminin5.In addition, we show that exogenous r-LEKTI blocks migration of OSC19-parental cells in a dose and time dependent manner. Microarray analysis identified 186 genes which are differentially regulated in both OSC19 LEKTI clones.mMP-14, KLK5, and ADAM8 are down regulated whilemMP-3, LEKTI, DSC2 and DSC3are up-regulated in OSC19 LEKTI clones. RT-PCR and Western blot results confirmed microarray results formMP-14 andmMP-3in OSC19 LEKTI clones. In addition we discover thatmMP-9 protein expression and pro-MMP-9 activity are severely reduced in LEKTI expressing clones as shown by WB and zymogram. Together, this work provides mechanistic insights into how loss of LEKTI protein expression promotes an invasive phenotype in HNSCC tumors.
Keywords: SPINK5, LEKTI, invasion, migration, adhesion, mMPs, HNSCC, KLK
LEKTI, lympho-epithelial kazal-type-inhibitor; SPINK5, serine protease inhibitor kazal-type 5; KLK, kallikreins; kDa, kilodaltons; RT-PCR, reverse-transcriptase polymerase chain reaction; SD, standard deviation of the mean
Lympho-epithelial kazal-type-inhibitor (LEKTI)1 was named by one of the original groups who cloned this protein’s gene to reflect the observed pattern of its expression in both epithelial tissue and leukocytes.1 It was later identified as the same gene as the defective gene in Netherton’s syndrome, SPINK5 (serine protease inhibitor Kazal-type 5).2 Netherton’s syndrome is a genetic disorder characterized by congenital ichthyosis, hair shaft abnormalities, immune deficiency, elevated immunoglobulin E (IgE) concentration, and failure to thrive.2–13 SPINK5 encodes the LEKTI protein which consists of 1064 amino acids organized into 15 potential inhibitory domains on the basis of the furin cleavage sites found within the full-length molecule. At the N-terminus is a secretory signal peptide sequence consisting of 22 amino acids.14 Two of the 15 LEKTI domains (domains 2 and 15) resemble typical Kazal-type serine proteinase inhibitors; the remaining 13 domains share partial homology to Kazal-type inhibitors but lack one of the three conserved Kazal-type disulfide bridges.15
We and others identified SPINK5 as one of the genes down regulated in head and neck squamous cell carcinoma (HNSCC).16,17 We cloned the cDNA encoding the 125-kDa isoform and established that recombinant pro-LEKTI is a potent inhibitor of multiple serine proteinases implicated in metastasis and angiogenesis. Moreover, rLEKTI did not inhibit the cysteine proteinase papain or cathepsin K, L, or S. We further showed that recombinant pro-LEKTI was very efficiently cleaved in vitro by furin into five major and thirteen minor proteolytic fragments.18 In the course of studies aimed at understanding the structure and function of some of these domains, we demonstrated that recombinant LEKTI6-9´ inhibited trypsin and subtilisin A but not plasmin, cathepsin G, or elastase.19 We also produced a battery of LEKTI monoclonal antibodies and demonstrated that several of these LEKTI antibodies including 1C11G6 reacted specifically with pro-LEKTI, LEKTI domains 1-6, 6-9, 9-12, and 12-15.20 We demonstrated that the N-terminal signal peptide is required for LEKTI import into the ER and ordered the cleavage products on the 125kDa pro-LEKTI from the amino- to carboxy-terminal as follows: 37-, 40-, and 60kDa.21 In our subsequent work, we characterized the interaction of two recombinant LEKTI domains 6-8 and 9-12 with recombinant rhK5 and recombinant rhK7.22 We showed that both fragments inhibited rhK5 similarly and established that LEKTI, at least in fragment form, is a potent inhibitor of rhK5 and that this protease may be a target of LEKTI in human skin. In our later studies we discovered that KLK5, KLK6, KLK13 and KLK14 were potently inhibited by rLEKTI (1-6), rLEKTI (6-9’) and rLEKTI (9-12).23,24 We also assessed the basis for phenotypic variations in patients with “mild”, “moderate” and “severe” NS.25 We observed that the magnitude of KLK activation correlated with both the barrier defect and clinical severity, and inversely with residual LEKTI expression and LEKTI co-localizes within the stratum corneum (SC) with kallikreins 5 and 7 and inhibits both KLKs. Recently, we demonstrated that caspase 14 is inhibited by full-length LEKTI and 5 recombinant fragments of LEKTI to varied extents.26
In the present study, we stably re-expressed LEKTI in HNSCC cells and evaluated the effects of LEKTI re-expression on cellular proliferation, morphology, adhesion, invasion and expression of key mMPs involved in tumor progression. LEKTI re-expression in OSC19 cells causes striking morphological changes, strongly enhances adhesion, markedly decreased migration and invasion. Stable re-expression of LEKTI in OSC19 cells resulted in markedly decreased levels of mMP-9 and mMP-14. Furthermore, these results demonstrate a novel negative regulatory role for LEKTI in modulating the production of keymMPs involved in ECM degradation and suggest that loss of LEKTI in HNSCC tumor cells could have a pivotal role in HNSCC progression.
Materials
The following reagents were obtained commercially as indicated: Human embryonic kidney cells (HEK 293T) (American Type Culture Collection, Manassas, VA); primary normal epidermal keratinocytes (HNEKs) and keratinocyte growth medium (Cambrex Biosciences, Walkersville, MD); OSC-19 from Dr. There OPTIMEM (Life Technologies, Rockville, MD); precast sodium dodecyl sulfate (SDS)-polyacrylamide gels, prestained markers, gelatin Zymogram gels (Bio-Rad Laboratories, Hercules, CA); nitrocellulose membrane (Schleicher & Schull BioScience, Keene, NH); YM3 Centriplus (Millipore, Bedford, MA); anti-LEKTI mAb1C11G6 (Zymed Laboratories, San Francisco, CA); collagens, I, III, and V, fibronectin, laminin-5, BSA, GAPDH, anti-β actin, anti-MMP-9, and anti-MMP–14 antibodies (Sigma-Aldrich, St. Louis, MO), horseradish peroxidase-conjugated goat-anti-mouse IgG (H+L) (Jackson ImmunoResearch Laboratories, West Grove, PA); lipofectamine 2000 and pcDNA3.1 (-) (Invitrogen, Carlsbad, CA); ECL kit (Amersham Bioscience Corporation, Piscataway, NJ); Kodak X-AR5 films (Eastman Kodak, Rochester, NY); restriction endonucleases and polymerase chain reaction reagents (New England Biolabs, Beverely, MA); BD BioCoat Tumor Invasion System (BD Biosciences); r-LEKTI is purified in our laboratory as described previously.18
Cell culture and transfections
The HNSCC cell lines (Tu 138 and JMAR) were established at The University of Texas M. D. Anderson Cancer Center. Dr. Tom Carey at the University of Michigan developed UMSCC1. A human oral squamous cell carcinoma cell strain, OSC19, was obtained from Dr. Theresa Whiteside and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 2mM glutamine, and antibiotics. HEK 293T were cultured in DMEM supplemented with 10% FBS and 2mM glutamine. HNEKs were cultured in keratinocyte growth medium containing low-calcium. All cells were cultured at 37°C in humidified incubator with 5% CO2 and 95% air. All cells were cultured at 37°C in humidified incubator with 5% CO2 and 95% air.
To clone the pro-LEKTI expression plasmid, 3.24Kb BamHI-KpnI fragment from LEKTI/pFASTBAC1, clone #4 was sub cloned into pcDNA 3.1. The pro-LEKTI expression plasmid encodes the entire full-length LEKTI polypeptide and has a hexahistidine tag at its C-terminus. 293T cells transfected with this construct expressed LEKTI. At 24h and 48h post transfection, LEKTI is detected within the cells and in the medium. In the medium LEKTI processed LEKTI fragments are also detected with LEKTI mAb 1C11G6. In order to delete the signal sequence, this clone is digested with NotI and EcoRI and ligated to a PCR fragment lacking this signal sequence. The pro-LEKTI-∆1-22 expression plasmid encodes the full-length LEKTI polypeptide without the N-terminus secretory signal sequence and has a hexahistidine tag at its C-terminus. When transfected into 293T cells, very little intracellular protein is observed. Following the verification of these expression clones, HNSCC cells were transfected with these constructs and stable clones were selected after G418 (0.5mg/ml) selection. We also performed transient transfection of OSC-19 and HEK 293T cells with these constructs. Cells were plated in 60×15-mm tissue culture dishes grown to 70% confluence, and transiently transfected with 2.0µg each of pro-LEKTI expression plasmid DNA or pro-LEKTI-∆1-22 expression plasmid DNA or control vector plasmid DNA using Lipofectamine 2000 according to the manufacturer’s instructions. Approximately 50% transfection efficiency was achieved as determined by transfection with a GFP control plasmid.
LEKTI secretion assays
Cells were washed twice in phosphate buffered saline (PBS) and resuspended in 4ml serum-free media for 24 hrs. The conditioned media was ten-fold concentrated by centrifugation using Millipore-YM3 Centriplus units (3,000 MWCO) and assayed for protein concentrations.27 To examine LEKTI expression in protein lysates, the adherent cells were trypsinized and solubilized in 100µl of ice-cold radio immunoprecipitation assay (RIPA) buffer containing 1% Nonidet P-40, 1.0% deoxycholate, 0.1% SDS, 50mM Tris-HCl, (pH 7.5), 150mM NaCl, 2mM EDTA and a mixture of protease inhibitors.
Western blot
Proteins or concentrated culture supernatants were mixed with 2×gel loading buffer (4% SDS; 20% glycerol; 120mM Tris-HCl, pH 6.8; 0.01% bromophenol blue; with or without 10 % β-mercaptoethanol), heated to 95°C for 5min, and resolved by SDS-PAGE (10% gel). Electrophoretic transfer of proteins from the polyacrylamide gel onto a nitrocellulose membrane (Schleicher & Schull BioScience, Inc., Keene, NH) was achieved by using amini-trans blot electrophoretic cell (Bio-Rad) at 25V for 16h at 4°C. After blocking the nitrocellulose membrane overnight at room temperature with 3% BSA, it was incubated for 2h at room temperature with primary and 1h with the secondary antibody. The immunoblot was visualized using the chemiluminescence-ECL substrate and exposed to X-ray Hyperfilm MP for 1-3min.
Adhesion assays
The 96-well high binding non- tissue culture plates were coated with equimolar amounts of type I, III, IV, and V collagens, fibronectin, and laminin-5, and vitronectin (100nM) for 3-4 hours at 37°C. Control wells received BSA alone. Wells were blocked with 5% BSA for 45minutes prior to use. HNSCC cells are added at 2X104 cells/well in serum-free DMEM. Cells were allowed to adhere at 37°C for 90minutes. Following washing, remaining adherent cells were fixed with 0.5% crystal violet in methanol/water. Background binding was assessed using coated wells but received no cells and subtracted from experimental values. The crystal violet incorporated into cell were collected in 1% SDS and quantified by measuring A590.
Migration and Invasion assays
Cell migration and invasion was determined using BD BioCoat Tumor Invasion System that consists of a 24-Mutliwell-insert plate in which a PET membrane (8µm pore size) has been coated without or with Matrigel. OSC19-parent, OSC19-Vector-1, OSC19-LEKTI-11, and OSC19-LEKTI-17 clones suspended in medium without serum were added to the upper wells of inserts, which were then were placed into lower wells containing NIH 3T3 supernatant containing 10% fetal calf serum as a chemo attractant and allowed to invade for 24h in a CO2 humidified incubator. To measure migration alone, parallel wells were set up with control inserts that lacked a Matrigel coating. Alternatively, OSC19-parent cells suspended in medium without serum and treated with recombinant pro-LEKTI were added to the upper wells of control inserts which were then were placed into lower wells containing NIH 3T3 supernatant containing 10% fetal calf serum as a chemo attractant and allowed to migrate for 24h in a CO2 humidified incubator. At the end of each assay, the lower sides of inserts containing migrated or invaded cells are stained and photographed.
Gelatin zymography
OSC19-parent, OSC19-Vector-1, OSC19-LEKTI-11, and OSC19-LEKTI-17 clones grown in DMEM media containing 10% fetal calf serum and 2mM glutamine were washed twice in phosphate buffered saline (PBS) and resuspended in 4ml of serum-free media for 24hrs. Thereafter, 1ml of supernatants was collected, centrifuged (300xg) for 10min to remove non-adherent cells and thereafter, supernatants were centrifuged a second time (5000xg) for 10min to remove cell debris and nuclei. Supernatants were concentrated (10X) by centrifugation using Millipore-YM3 Centriplus units (3,000 MWCO). 5µl concentrated supernatants are mixed with Zymogram sample buffer in the absence of reducing agents and electrophoresed through 12% polyacrylamidegels containing 0.1% (w/v). Electrophoresis was carried out at 125 V. After electrophoresis, the gel was washed twice with 100ml of 2.5% (v/v) Triton X-100 at 22°C for 30min to remove SDS, and three times for 10min with H2O to remove Triton X-100. The gel was incubated in 50mM Tris-HCl, 0.2 M NaCl, 20mM CaCl2, pH 7.4 at 37 C for 12h, stained over night with Coomassie Brilliant Blue R-250 0.5% (w/v) in 45% (v/v) methanol-10% (v/v) acetic acid and destained in the same solution without dye. The location of gelatinolytic activity is visualized as a clear band on the uniformly stained background.
Northern blot and Real time PCR
Total RNA isolation, Northern blot and Real-time PCR were performed as described earlier.28 Total RNA was prepared using TriZol reagent (Invitrogen) according to the manufacturer’s instructions. For Northern blot, 20µg total RNA was applied to a 1% formaldehyde agarose gel. After transferring RNA to Hybond-Nþ membrane (Amersham), the membrane and the filter were hybridized with 32P-r-human LEKTI or 32P-GAPDH. For Real-time PCR, 2µg total RNA were reverse transcribed (RT) by Superscript II (Life Technologies) in a 25µl total reaction volume containing RT buffer, random hexamers, dNTP, and RNase inhibitor (Roche Applied Science, Indianapolis, IN). Real-time PCR was performed in a 25µl total reaction volume containing 1µ of 1:10 diluted cDNA obtained from RT reaction, 12.5µl of TaqMan Universal PCR Master Mix without AmpErase UNG, and 1.25µl of specific primers for each gene on ABI Prism 7900HT (kindly provided by Dr. Adel El-Naggar from the Department of Pathology, M. D. Anderson Cancer Center). As a control, 18S primers were used, and cDNA was diluted to 1:500. Serial dilutions of the standard templates were also used for parallel amplifications. The threshold cycles (Ct) were calculated with ABI Prism 7900HT SDS software (Applied Biosystems). The quantities of samples were determined from the standard curves. Levels ofmMP-3, mMP-9 andmMP-19 mRNA were normalized to those of 18S in each sample. For statistical analysis, the Tukey HSD analysis of variance post hoc test was used as a univariate test for significant differences between the ratio means and the P value was determined. A P value of 0.05 or less was considered significant.
Microarray analysis
Total cellular RNA isolation, cDNA preparation, and microarray analysis were performed as described previously.28,29 Briefly, hybridization to microarrays was performed using human oligonucleotide–spotted glass array with 18,861 60-mer oligos and controls produced in the Wiegand Radiation Oncology Microarray Core Facility at our institution. Hybridization was carried out for 16h at 50°C. Scanned images were quantified in Array Vision (Imaging Research, Inc., St. Catherine’s, Ontario, Canada). Measurements were recorded for spot intensity, local background intensity, and signal-to-noise ratio. Spot intensity was computed as the integrated absorbance or volume in a fixed-size circle. Background intensity was computed as the median pixel value in four diamond-shaped regions at the corners of each spot. The signal-to-noise ratio was computed by dividing the background-corrected intensity by the SD of the background pixels. Quantified array data were imported into S-Plus software (Insightful Corp., Seattle, WA) for analysis. Background-corrected intensities were globally rescaled to set the 75th percentile in each channel equal to 1,024. Rescaled intensities of <128 were replaced by the threshold value; this threshold was chosen to lie just below the smallest intensity of any spot with a signal-to-noise ratio >1. Next, intensities were transformed by computing the base 2 logarithm. Finally, the log-transformed spot intensities were normalized using robust local regression. A spot was identified as differentially expressed if the mean intensity in the two channels exceeded 512 and the estimated change exceeded 2.5-fold or if the mean intensity in the two channels exceeded 256 and the estimated change exceeded 4.5-fold. The normalized data was logarithm transformed to base 2 and the mean data of the replicates was determined. The log ratio values were calculated for the clones 11 and 17 and corresponding paired vector controls. Differentially regulated mRNA between the 2 samples was identified using a paired t test. We included the complete microarray dataset as a supplementary Microsoft Excel file and also in the process of depositing our dataset to Array Express database soon.
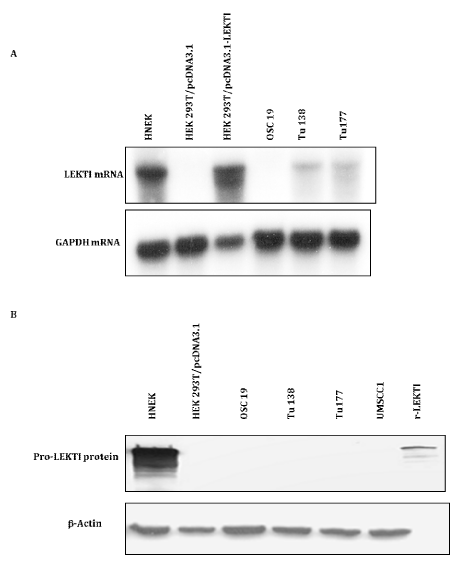
Analysis of LEKTI mRNA and LEKTI protein expression in HNSCC tumor lines
Using Northern blotting, we analyzed the expression of LEKTI mRNA in HNEK, HEK 293T-vector transfect ant, HEK 293T-LEKTI transfect ant, and HNSCC OSC19, Tu138 and Tu177 tumor lines. Northern blot analysis showed that a 3.75-kb mRNA band was detected in HNEK and 293T cells transiently transfected with pro-LEKTI expression plasmid. This transcript is not detectable in 293T-vector transfectant and OSC19 cells but present in reduced levels in Tu177 and Tu138 cell lines. The results were consistent with the patterns of LEKTI mRNA expression previously reported for normal oral epithelium and several HNSCC cell lines including Tu138 and Tu 177 (Figure 1A).16 Using Western blotting, we analyzed the expression of native LEKTI protein by using anti-LEKTI mAb 1C11G6 and protein lysates from HNEK, HEK 293T-vector transfectant, and HNSCC OSC19, Tu138, Tu177, and UMSCC1 tumor lines. Anti-LEKTI antibody 1C11G6 recognized a major protein of ∼125kDa and amine or protein of 110kDa in HNEKs indicating robust expression of LEKTI protein; in contrast both bands were absent in all four HNSCC tumor lines (Figure 1B). The endogenously expressed 125kDa LEKTI found in HNEKs was identical to r-pro-LEKTI expressed and purified from insect cells.18 Protein lysates from HEK 293T cell transfected with control vector showed no endogenous LEKTI expression. This Western blot signal was completely absent when a preserum was used. The results were consistent with the patterns of LEKTI protein expression reported for normal kidney and colon tissues.30
Stable LEKTI re-expression inhibits matrigel migration and invasion of OSC19
To understand the impact of LEKTI re-expression in HNSCC cells, we cloned a native LEKTI cDNA fragment encoding pro-LEKTI protein into the expression vector pcDNA3.1, transfected into OSC19 and UMSCC1 cells and isolated several stable clones after G418 (0.5mg/ml) selection for almost a month. We selected one OSC19-vector clone 1; two OSC19-LEKTI clones 11 and 17 and one UMSCC1 clone 2 for further studies. We examined the cell lysates and conditioned medium from these clones for the expression of recombinant and for the presence of processed LEKTI fragments by Western blot assays (Figure 2A). Stable expression of recombinant LEKTI in these three clones is processed and secreted into the medium similar to what we reported for HNEK cells.21 We recently showed that native pro-LEKTI in HNEK cells is processed and secreted into the medium by an ER/Golgi-dependent pathway. Furthermore, we demonstrated that endogenous furin plays a pivotal role in the processing pro-LEKTI. Now, we show that stably expressed recombinant LEKTI in OSC19 and UMSCC1cells is also similarly processed and secreted into the medium.
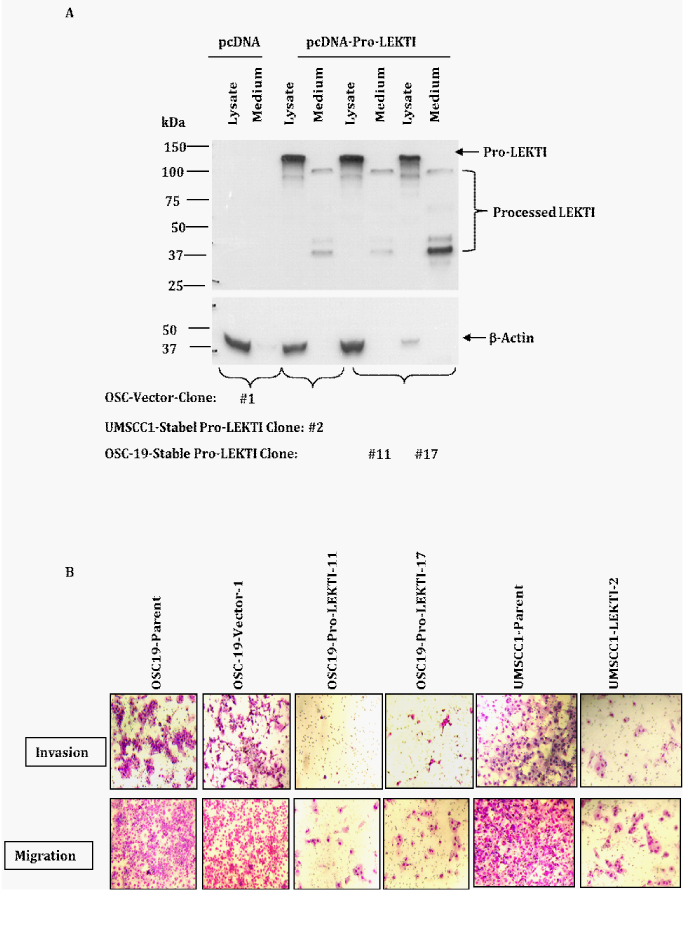
In our pilot studies we noticed no migration of any of our HNSCC lines in conditions in which no chemo attractant was added. Hence, we compared the chemotactic migration of various HNSCC tumor lines in response to 5% FCS or NIH3T3 supernatant in an uncoated control insert plate with a PET membrane containing 8micron pores. We found that NIH 3T3 supernatant proved a more optimal chemo attractant with a 30% increase in cell migration compared to 5% FBS. In addition, we observed a 2- to 3.5-fold increase in the migration of OSC19 compared with UMSCC1. On the basis of these results, we used the NIH 3T3 supernatant as a chemo attractant in our subsequent assays. In each of two LEKTI expressing clones of OSC-19, invasion was dramatically reduced compared to parental and vector cells (Figure 2B). Likewise, in LEKTI expressing clones of UMSCC2 invasion were significantly reduced compared to parental cells. We found more number of OSC19 parental and vector cells and UMSCC1 parental cells stained under migration than under invasion assays. Surprisingly, in each of two LEKTI expressing clones of OSC19 and one LEKTI expressing clones of UMSCC1, migration also was dramatically reduced compared to parental and vector cells. On the basis of these results, we conclude that LEKTI in tumor cells most likely functions in the extracellular tumor microenvironment and furthermore the levels of LEKTI in LEKTI transfected HNSCC cell model are comparable with those existing naturally. These data suggest that stable LEKTI expression results in inhibition of migration and invasion of HNSCC tumor lines in vitro.
Transient LEKTI re-expression inhibits matrigel migration of OSC19
To determine the effect of LEKTI re-expression on cellular migration, we transiently transfected OSC19 cells with pro-LEKTI or pro-LEKTI deleted for the N-terminal secretory sequence. Using these transfectants we first characterized LEKTI expression and then performed migration assays. We observed that the expression of carboxy-terminal hexa-histidine tagged pro-LEKTI with it signal sequence and its processed LEKTI fragments was abundant in the cell lysates and supernatant as shown by its immuno reactivity with LEKTI mAb (Figure 3A). In contrast, deletion of the signal peptide resulted in markedly low levels of pro-LEKTI-∆1-22 in the soluble fraction of cell lysates (Figure 3A), and we could not detect any processed LEKTI fragments in the conditioned medium (Figure 3A). These results are in agreement with our previous studies where we demonstrated that pro-LEKTI is processed and secreted into the medium in HEK 293T cells transiently transfected with the pro-LEKTI expression plasmid whereas the level of pro-LEKTI-∆1-22 in lysates and medium went down dramatically.21
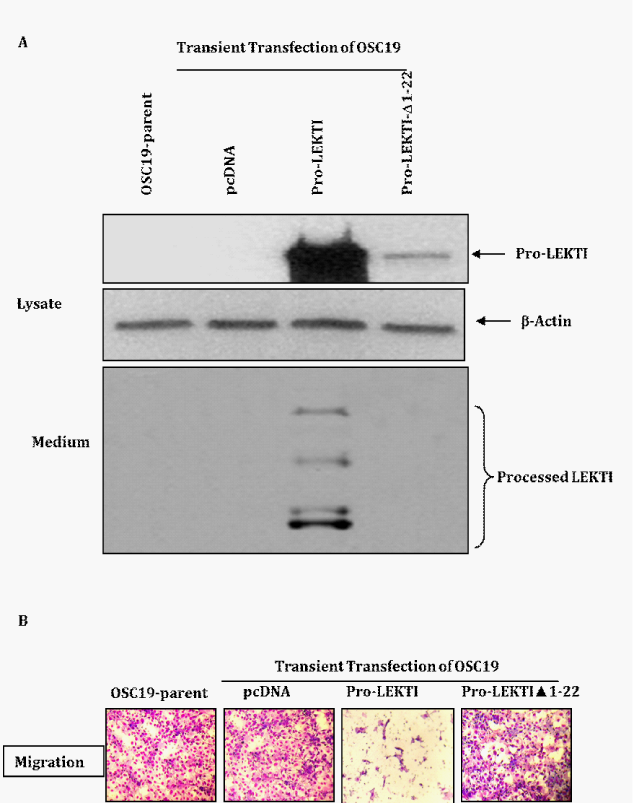
The migration of OSC19 cells transfected with empty vector was comparable to their un-transfected OSC19 parental cells (Figure 3B). In contrast, the migration of OSC19 Pro-LEKTI transfectent through Matrigel was inhibited by more than 95% (P=0.001). On the other hand, the migration of pro-LEKTI-∆1-22 transfectant through Matrigel was not affected and it is comparable to OSC19 cells transfected with empty vector and un-transfected OSC19 parental cells. On the basis of these results we conclude that LEKTI protein is directly responsible for the inhibition of OSC19 migration and also functional LEKTI is required to exert its inhibitory effect on migration.
Exogenous r-LEKTI inhibits matrigel invasion and migration of OSC19
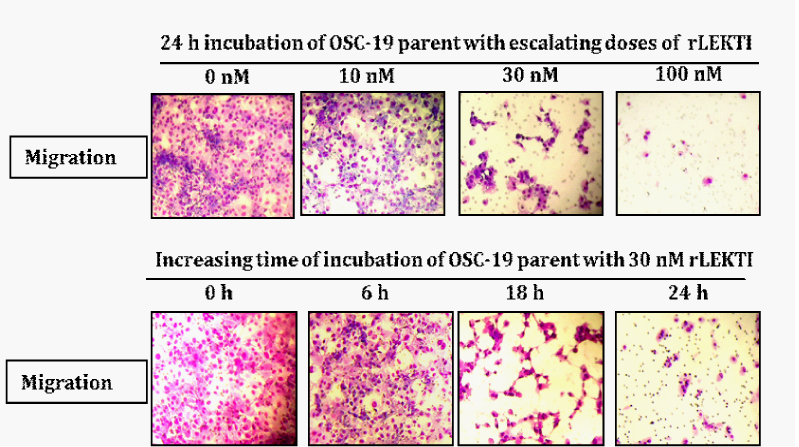
Next, we tested if exogenous r-LEKTI can elicit the inhibition of migration of OSC parental cells. Recombinant pro-LEKTI was expressed and purified from insect cells as described previously.18,23,31 OSC19 parental cells are treated without r-LEKTI or with 10nM, 30nM, and 100nM r-LEKTI for 24h or with 30nM r-LEKTI for 0h, 6h, 18h, and 24h and then cells were allowed to migrate for 24h with control inserts that lacked a Matrigel coating in a CO2 humidified incubator. Migration assays show that r-LEKTI inhibits the migration of OSC19 cells in a dose and time dependent fashion (Figure 4). No inhibition of migration was observed after 6 h incubation with 30nM r-LEKTI but 18h incubation resulted in substantial inhibition and 24h incubation showed almost a total inhibition.
LEKTI re-expression enhances adhesion and regulates the morphology of OSC19
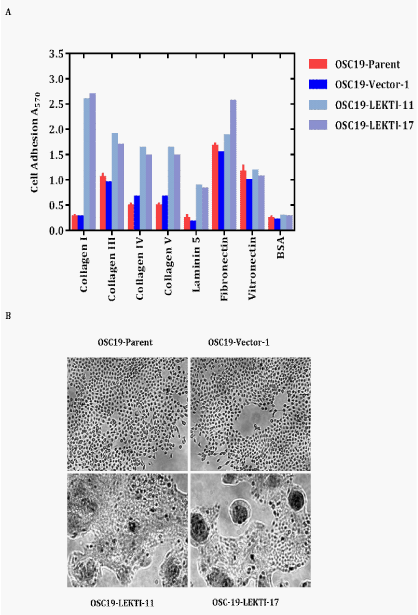
We observed that both OSC19 LEKTI clones required more time for trypsinization when passaging the cells. To test this observation and determine the adhesion of cells to extracellular matrix constituents in relation to LEKTI expression, we performed assays of OSC19 cell adhesion to collagen type I, collagen type III, collagen type IV, collagen type V, laminin-5, fibronectin, and vitronectin. After 3h of adhesion, each of two LEKTI-expressing clones demonstrated increased adhesion to collagen I, III, IV, IV, laminin-5, and fibronectin compared to parental and vector cells (Figure 5A). The increase in adhesion of OSC19-LEKTI clones to collagen I, III, IV, IV, laminin-5, and fibronectin was 1300, 100, 150, 200, 300 and 40% respectively. In contrast, when plated on vitronectin, both control and LEKTI stable clones showed a similar extent of adhesion. Additionally, the morphology of LEKTI-expressing OSC19 clones was drastically altered from the parental and vector cells. After 4days of culture in normal tissue culture plates, the parental and vector cells formed thin and spreading stellate shapes with numerous processes (Figure 5B). In contrast, each of two LEKTI-expressing clones of OSC19 formed polygonal shapes and aggregated into compacts clumps. On the basis of these results, we conclude that re-expressed LEKTI in HNSCC cells lead to a cell type specific increase in adhesion onto specific extracellular matrix substrates. These observations suggest that LEKTI expression regulates cell morphology to result in a more differentiated phenotype resembling the architecture of squamous epithelium.
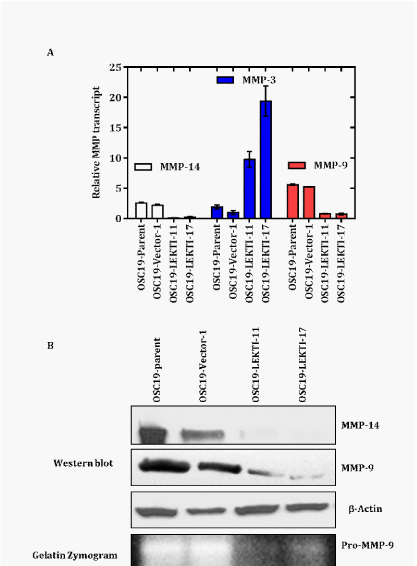
LEKTI re-expression negatively regulates expression ofmMP-9 and -14
Microarray analysis identified 186 genes which are differentially regulated between LEKTI clones in comparison with the vector transfected OSC19. Among them,mMP-14,mMP-8, KLK5, and ADAM8 are down regulated andmMP-3, LEKTI, DSC2 and DSC3 are up-regulated in LEKTI clones (Table 1) of the four classes of proteolytic enzymes, which include mMPs, serine proteinases, cysteine proteinases, and aspartate proteinases, them MPs are most important to the process of metastasis.32,33 To examine the expression of mMPs in relation to LEKTI expression we performed analysis of mMPs in OSC19 parental, vector, and LEKTI- expressing clones 11 and 17. By real-time PCR, each LEKTI-expressing clone showed a decrease in mMP-14 and an increase inmMP-3 transcript (Figure 6A). These results confirmed our microarray results on these two mMPs (Table 2). In addition, we found out that each LEKTI-expressing clone showed a decrease inmMP-9 mRNA expression relative to parental and vector cells (Figure 6A). Consistent with down regulation ofmMP-9 and mMP-14 mRNA expression, each LEKTI-expressing clone showed dramatic reduction in mMP-9 and –14-protein level relative to parental and vector cells (Figure 6B). We confirmed this finding by zymogram showing a decrease in mMP-9 protein activity levels in response to LEKTI expression (Figure 6B).
Spot Location |
UGRepAcc |
Symbol |
Name |
Clone |
Clone |
Up regulation |
|||
Foldchange_clone11 |
Foldchange_clone17 |
||||||||
C-6:23-8 |
NM_001512 |
GSTA4 |
glutathione S-transferase A4 |
15.86 |
14.37 |
59575.04 |
21128.97 |
||
A-8:12-11 |
NM_002373 |
MAP1A |
microtubule-associated protein 1A |
15.86 |
14.99 |
59300.62 |
32530.79 |
||
B-3:17-5 |
NM_006536 |
CLCA2 |
chloride channel, calcium activated, family member 2 |
14.95 |
2.02 |
31659.09 |
4.04 |
||
D-7:16-2 |
AL050367 |
LOC221061 |
hypothetical protein LOC221061 |
14.30 |
14.73 |
20215.06 |
27092.83 |
||
B-2:6-14 |
NM_018372 |
RIF1 |
receptor-interacting factor 1 |
14.15 |
4.94 |
18239.04 |
30.70 |
||
A-8:4-3 |
XM_290809 |
TAF4B |
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa |
13.92 |
15.21 |
15523.79 |
37944.50 |
||
B-6:1-23 |
NM_006180 |
NTRK2 |
neurotrophic tyrosine kinase, receptor, type 2 |
13.87 |
4.72 |
14952.63 |
26.31 |
||
A-5:16-1 |
NM_025247 |
ALDH2 |
aldehyde dehydrogenase 2 family (mitochondrial) |
13.45 |
3.30 |
11189.55 |
9.84 |
||
A-6:20-14 |
NM_001449 |
FHL1 |
four and a half LIM domains 1 |
13.34 |
14.21 |
10337.80 |
18959.51 |
||
C-7:24-1 |
NM_000691 |
ALDH3A1 |
aldehyde dehydrogenase 3 family, memberA1 |
9.53 |
6.70 |
737.55 |
103.98 |
||
D-2:12-5 |
NM_002421 |
MMP1 |
matrix metalloproteinase 1 (interstitial collagenase) |
8.14 |
14.06 |
281.19 |
17059.21 |
||
A-1:2-10 |
NM_002638 |
PI3 |
protease inhibitor 3, skin-derived (SKALP) |
7.84 |
3.57 |
229.76 |
11.84 |
||
B-2:13-12 |
NM_002964 |
S100A8 |
S100 calcium binding protein A8 (calgranulin A) |
6.32 |
5.85 |
79.70 |
57.61 |
||
D-3:7-14 |
NM_004561 |
OVOL1 |
ovo-like 1(Drosophila) |
6.10 |
3.79 |
68.59 |
13.83 |
||
C-4:25-15 |
NM_006783 |
GJB6 |
gap junction protein, beta 6 (connexin 30) |
6.07 |
14.86 |
67.11 |
29790.05 |
||
A-8:24-15 |
NM_014220 |
TM4SF1 |
transmembrane 4 superfamily member 1 |
5.53 |
5.90 |
46.05 |
59.70 |
||
D-5:17-6 |
NM_006227 |
PLTP |
phospholipid transfer protein |
5.38 |
5.30 |
41.55 |
39.44 |
||
C-2:4-11 |
NM_002272 |
KRT4 |
keratin 4 |
5.37 |
2.57 |
41.47 |
5.92 |
||
B-2:11-1 |
NM_005329 |
HAS3 |
hyaluronan synthase 3 |
5.34 |
4.21 |
40.39 |
18.45 |
||
A-3:25-9 |
NM_153490 |
KRT13 |
keratin 13 |
5.32 |
3.73 |
40.06 |
13.27 |
||
B-1:15-1 |
NM_022746 |
FLJ22390 |
hypothetical protein FLJ22390 |
5.29 |
2.64 |
39.20 |
6.24 |
||
D-3:8-6 |
NM_001353 |
AKR1C1 |
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase) |
5.26 |
6.41 |
38.19 |
85.32 |
||
B-1:23-10 |
NM_003914 |
CCNA1 |
cyclin A1 |
5.23 |
5.48 |
37.61 |
44.58 |
||
C-7:1-3 |
NM_005986 |
SOX1 |
SRY (sex determining region Y)-box 1 |
5.20 |
5.19 |
36.72 |
36.40 |
||
D-5:15-6 |
|
|
|
5.18 |
6.15 |
36.18 |
70.99 |
||
B-3:19-5 |
NM_002422 |
MMP3 |
matrix metalloproteinase 3 (stromelysin 1, progelatinase) |
5.04 |
5.65 |
33.01 |
50.37 |
||
D-5:18-3 |
NM_181353 |
ID1 |
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein |
4.90 |
4.26 |
29.80 |
19.13 |
||
B-2:19-14 |
NM_003125 |
SPRR1B |
small proline-rich protein 1B (cornifin) |
4.61 |
2.79 |
24.46 |
6.94 |
||
C-1:18-14 |
NM_017459 |
MFAP2 |
microfibrillar-associated protein 2 |
4.57 |
13.81 |
23.77 |
14373.79 |
||
D-3:18-8 |
NM_007366 |
PLA2R1 |
phospholipase A2 receptor 1, 180kDa |
4.51 |
4.30 |
22.74 |
19.64 |
||
D-6:21-8 |
NM_000584 |
IL8 |
interleukin 8 |
4.42 |
2.03 |
21.34 |
4.08 |
||
A-6:12-11 |
NM_001978 |
EPB49 |
erythrocyte membrane protein band 4.9 (dematin) |
4.21 |
3.03 |
18.56 |
8.16 |
||
C-7:13-8 |
NM_024829 |
FLJ22662 |
hypothetical protein FLJ22662 |
4.10 |
2.68 |
17.20 |
6.40 |
||
C-3:1-7 |
NM_145791 |
MGST1 |
microsomal glutathione S-transferase 1 |
4.06 |
5.85 |
16.67 |
57.66 |
||
D-7:5-9 |
NM_014656 |
KIAA0040 |
KIAA0040 gene product |
3.92 |
2.46 |
15.12 |
5.51 |
||
B-6:14-2 |
NM_001964 |
EGR1 |
early growth response 1 |
3.88 |
4.25 |
14.75 |
19.01 |
||
D-7:13-24 |
NM_144665 |
SESN3 |
sestrin 3 |
3.85 |
5.80 |
14.41 |
55.80 |
||
C-6:14-23 |
AK001903 |
|
CDNA FLJ11041 fis, clone PLACE1004405 |
3.77 |
4.94 |
13.67 |
30.61 |
||
B-1:15-6 |
NM_002130 |
HMGCS1 |
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 (soluble) |
3.75 |
2.73 |
13.48 |
6.62 |
||
C-4:22-8 |
NM_006846 |
SPINK5 |
serine protease inhibitor, Kazal type, 5 |
3.73 |
14.85 |
13.31 |
29489.80 |
||
C-1:22-9 |
NM_002275 |
KRT15 |
keratin 15 |
3.57 |
2.40 |
11.88 |
5.28 |
||
D-7:20-3 |
NM_004473 |
FOXE1 |
forkhead box E1 (thyroid transcription factor 2) |
3.54 |
3.21 |
11.66 |
9.22 |
||
D-3:2-18 |
NM_032508 |
FAM11A |
family with sequence similarity 11, member A |
3.39 |
4.87 |
10.47 |
29.26 |
||
C-4:17-23 |
AK026158 |
LOC348938 |
hypothetical protein LOC348938 |
3.37 |
4.67 |
10.37 |
25.42 |
||
B-5:5-1 |
NM_007286 |
SYNPO |
synaptopodin |
3.37 |
3.52 |
10.37 |
11.46 |
||
B-7:13-13 |
NM_000299 |
PKP1 |
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) |
3.36 |
2.90 |
10.24 |
7.48 |
||
C-5:3-4 |
NM_001452 |
FOXF2 |
forkhead box F2 |
3.35 |
2.70 |
10.22 |
6.50 |
||
D-7:16-23 |
NM_031455 |
CCDC3 |
coiled-coil domain containing 3 |
3.33 |
3.36 |
10.03 |
10.26 |
||
D-8:25-5 |
NM_012244 |
SLC7A8 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 |
3.23 |
5.26 |
9.36 |
38.32 |
||
B-2:24-8 |
NM_002996 |
CX3CL1 |
chemokine (C-X3-C motif) ligand 1 |
3.22 |
6.33 |
9.29 |
80.62 |
||
B-6:6-19 |
NM_004988 |
MAGEA1 |
melanoma antigen, family A, 1 (directs expression of antigen MZ2-E) |
3.13 |
4.28 |
8.74 |
19.45 |
||
D-7:24-15 |
NM_018950 |
HLA-F |
major histocompatibility complex, class I, F |
3.10 |
2.71 |
8.60 |
6.52 |
||
D-2:13-16 |
AF001893 |
|
MRNA; cDNA DKFZp686L01105 (from clone DKFZp686L01105) |
3.08 |
2.59 |
8.43 |
6.04 |
||
A-7:8-7 |
NM_194298 |
SLC16A9 |
solute carrier family 16 (monocarboxylic acid transporters), member 9 |
3.07 |
3.48 |
8.41 |
11.18 |
||
B-3:22-3 |
NM_016307 |
PRRX2 |
paired related homeobox 2 |
3.05 |
2.72 |
8.26 |
6.57 |
||
D-5:11-12 |
NM_001775 |
CD38 |
CD38 antigen (p45) |
3.04 |
2.37 |
8.25 |
5.16 |
||
C-8:25-6 |
NM_001360 |
DHCR7 |
7-dehydrocholesterol reductase |
3.00 |
2.53 |
8.01 |
5.80 |
||
D-5:24-12 |
NM_005130 |
HBP17 |
heparin-binding growth factor binding protein |
3.00 |
2.25 |
8.00 |
4.77 |
||
C-2:11-23 |
BX640887 |
|
CDNA clone IMAGE:3880075, partial cds |
2.98 |
3.23 |
7.88 |
9.40 |
||
D-1:7-6 |
NM_001823 |
CKB |
creatine kinase, brain |
2.95 |
4.54 |
7.73 |
23.24 |
||
D-4:21-6 |
NM_002083 |
GPX2 |
glutathione peroxidase 2 (gastrointestinal) |
2.88 |
3.50 |
7.38 |
11.31 |
||
B-6:15-10 |
NM_006763 |
BTG2 |
BTG family, member 2 |
2.82 |
2.25 |
7.05 |
4.77 |
||
B-5:14-12 |
NM_003028 |
SHB |
SHB (Src homology 2 domain containing) adaptor protein B |
2.80 |
16.66 |
6.97 |
103825.37 |
||
B-5:17-11 |
NM_004949 |
DSC2 |
desmocollin 2 |
2.78 |
2.04 |
6.86 |
4.12 |
||
B-5:19-11 |
NM_002214 |
ITGB8 |
integrin, beta 8 |
2.73 |
3.22 |
6.65 |
9.32 |
||
C-7:19-9 |
NM_000422 |
KRT17 |
keratin 17 |
2.73 |
2.91 |
6.61 |
7.51 |
||
A-5:13-16 |
NM_000692 |
ALDH1B1 |
aldehyde dehydrogenase 1 family, member B1 |
2.61 |
2.33 |
6.12 |
5.03 |
||
D-2:12-17 |
NM_032333 |
MGC4248 |
hypothetical protein MGC4248 |
2.59 |
2.35 |
6.04 |
5.09 |
||
A-1:15-11 |
NM_005901 |
MADH2 |
MAD, mothers against decapentaplegic homolog 2 (Drosophila) |
2.59 |
4.06 |
6.02 |
16.71 |
||
D-8:16-14 |
NM_000852 |
GSTP1 |
glutathione S-transferase pi |
2.58 |
2.27 |
5.98 |
4.82 |
||
C-4:23-20 |
AK000090 |
|
CDNA FLJ20083 fis, clone COL03440 |
2.55 |
2.84 |
5.85 |
7.14 |
||
C-4:24-20 |
AK000794 |
|
CDNA FLJ20787 fis, clone COL02178 |
2.46 |
3.16 |
5.51 |
8.95 |
||
C-3:18-14 |
NM_003480 |
MAGP2 |
Microfibril-associated glycoprotein-2 |
2.43 |
6.55 |
5.37 |
93.83 |
||
D-3:22-7 |
NM_006822 |
RAB40B |
RAB40B, member RAS oncogene family |
2.39 |
2.64 |
5.25 |
6.22 |
||
B-5:16-8 |
|
|
|
2.38 |
4.25 |
5.21 |
19.02 |
||
A-8:16-9 |
NM_006096 |
NDRG1 |
N-myc downstream regulated gene 1 |
2.31 |
2.61 |
4.95 |
6.10 |
||
C-1:12-10 |
NM_004431 |
EPHA2 |
EphA2 |
2.27 |
2.30 |
4.83 |
4.94 |
||
D-6:8-3 |
NM_018555 |
ZNF331 |
zinc finger protein 331 |
2.27 |
3.64 |
4.81 |
12.44 |
||
B-7:19-6 |
NM_014427 |
CPNE7 |
copine VII |
2.26 |
2.79 |
4.80 |
6.90 |
||
C-2:8-7 |
XM_370652 |
DNCH2 |
dynein, cytoplasmic, heavy polypeptide 2 |
2.26 |
2.04 |
4.79 |
4.11 |
||
A-1:25-13 |
NM_002820 |
PTHLH |
parathyroid hormone-like hormone |
2.26 |
3.14 |
4.78 |
8.82 |
||
C-4:18-11 |
NM_018103 |
LRRC5 |
leucine rich repeat containing 5 |
2.23 |
3.37 |
4.70 |
10.32 |
||
A-8:13-1 |
|
|
|
2.21 |
2.60 |
4.62 |
6.07 |
||
D-7:21-1 |
NM_000104 |
CYP1B1 |
cytochrome P450, family 1, subfamily B, polypeptide 1 |
2.21 |
2.32 |
4.61 |
5.00 |
||
A-1:20-17 |
NM_052932 |
PORIMIN |
pro-oncosis receptor inducing membrane injury gene |
2.18 |
2.04 |
4.53 |
4.12 |
||
C-2:1-3 |
NM_138281 |
DLX4 |
distal-less homeobox 4 |
2.17 |
3.85 |
4.50 |
14.41 |
||
D-4:19-17 |
AK024449 |
PP2135 |
PP2135 protein |
2.16 |
3.26 |
4.48 |
9.61 |
||
D-1:15-13 |
NM_001657 |
AREG |
amphiregulin (schwannoma-derived growth factor) |
2.16 |
2.43 |
4.47 |
5.37 |
||
D-1:13-2 |
NM_005642 |
TAF7 |
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
2.16 |
2.30 |
4.46 |
4.92 |
||
B-3:19-12 |
NM_000597 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
2.16 |
2.24 |
4.45 |
4.72 |
||
C-1:1-18 |
BQ431041 |
|
LOC388279 (LOC388279), mRNA |
2.15 |
2.72 |
4.44 |
6.60 |
||
B-2:17-21 |
NM_007006 |
CPSF5 |
cleavage and polyadenylation specific factor 5, 25 kDa |
2.14 |
2.36 |
4.41 |
5.13 |
||
B-4:11-17 |
NM_006096 |
NDRG1 |
N-myc downstream regulated gene 1 |
2.14 |
2.39 |
4.41 |
5.24 |
||
D-6:15-7 |
|
|
|
2.09 |
2.05 |
4.24 |
4.15 |
||
A-1:17-3 |
NM_016265 |
ZNF325 |
zinc finger protein 325 |
2.08 |
2.29 |
4.24 |
4.89 |
||
B-4:16-11 |
NM_024423 |
DSC3 |
desmocollin 3 |
2.08 |
2.43 |
4.23 |
5.39 |
||
C-4:17-7 |
NM_006598 |
SLC12A7 |
solute carrier family 12 (potassium/chloride transporters), member 7 |
2.03 |
2.62 |
4.08 |
6.13 |
||
D-5:8-6 |
NM_031220 |
PITPNM3 |
PITPNM family member 3 |
2.01 |
2.94 |
4.02 |
7.70 |
||
A-7:1-16 |
NM_015714 |
G0S2 |
putative lymphocyte G0/G1 switch gene |
-2.02 |
-2.59 |
0.25 |
0.17 |
4.067469826 |
6.02797569 |
D-4:7-17 |
NM_000202 |
IDS |
iduronate 2-sulfatase (Hunter syndrome) |
-2.03 |
-2.75 |
0.25 |
0.15 |
4.077710921 |
6.75034936 |
D-3:6-9 |
NM_000202 |
IDS |
iduronate 2-sulfatase (Hunter syndrome) |
-2.04 |
-2.87 |
0.24 |
0.14 |
4.101875487 |
7.29841054 |
C-5:25-16 |
NM_138768 |
MYEOV |
myeloma overexpressed gene (in a subset of t(11;14) positive multiple myelomas) |
-2.05 |
-2.49 |
0.24 |
0.18 |
4.144497434 |
5.62253016 |
D-3:23-9 |
NM_018222 |
PARVA |
parvin, alpha |
-2.05 |
-2.56 |
0.24 |
0.17 |
4.145801706 |
5.88473227 |
B-7:4-23 |
NM_182507 |
LOC144501 |
hypothetical protein LOC144501 |
-2.07 |
-4.11 |
0.24 |
0.06 |
4.208758818 |
17.3227835 |
C-8:22-13 |
NM_020799 |
AMSH-LP |
associated molecule with the SH3 domain of STAM (AMSH) like protein |
-2.08 |
-2.34 |
0.24 |
0.20 |
4.214203909 |
5.04987403 |
A-5:10-10 |
NM_003087 |
SNCG |
synuclein, gamma (breast cancer-specific protein 1) |
-2.11 |
-2.18 |
0.23 |
0.22 |
4.30472467 |
4.53020489 |
D-7:12-1 |
NM_014297 |
ETHE1 |
ethylmalonic encephalopathy 1 |
-2.14 |
-2.15 |
0.23 |
0.23 |
4.419559792 |
4.43492191 |
A-2:18-8 |
NM_002543 |
OLR1 |
oxidised low density lipoprotein (lectin-like) receptor 1 |
-2.18 |
-4.00 |
0.22 |
0.06 |
4.518721967 |
15.9888148 |
D-3:16-8 |
NM_000963 |
PTGS2 |
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
-2.18 |
-2.20 |
0.22 |
0.22 |
4.533257978 |
4.60992803 |
C-8:1-10 |
NM_022481 |
ARAP3 |
ARF-GAP, RHO-GAP, ankyrin repeat and plekstrin homology domains-containing protein 3 |
-2.18 |
-2.59 |
0.22 |
0.17 |
4.542590739 |
6.02919293 |
C-7:5-7 |
NM_023927 |
NS3TP2 |
HCV NS3-transactivated protein 2 |
-2.19 |
-3.26 |
0.22 |
0.10 |
4.566722537 |
9.58576143 |
D-5:12-13 |
NM_000024 |
ADRB2 |
adrenergic, beta-2-, receptor, surface |
-2.22 |
-2.05 |
0.21 |
0.24 |
4.674568202 |
4.13068734 |
D-4:19-2 |
NM_031283 |
TCF7L1 |
transcription factor 7-like 1 (T-cell specific, HMG-box) |
-2.25 |
-3.09 |
0.21 |
0.12 |
4.77061901 |
8.52709149 |
B-7:5-9 |
NM_000916 |
OXTR |
oxytocin receptor |
-2.29 |
-2.33 |
0.20 |
0.20 |
4.890803849 |
5.0310331 |
A-7:6-15 |
NM_003289 |
TPM2 |
tropomyosin 2 (beta) |
-2.32 |
-2.76 |
0.20 |
0.15 |
5.008541393 |
6.78462145 |
B-2:12-7 |
NM_015994 |
ATP6V1D |
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
-2.33 |
-2.16 |
0.20 |
0.22 |
5.025197605 |
4.46298352 |
D-3:18-11 |
NM_000610 |
CD44 |
CD44 antigen (homing function and Indian blood group system) |
-2.34 |
-2.76 |
0.20 |
0.15 |
5.056402704 |
6.77232189 |
C-3:12-5 |
NM_014256 |
B3GNT3 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
-2.35 |
-2.53 |
0.20 |
0.17 |
5.084537435 |
5.7804644 |
A-5:3-14 |
NM_005096 |
ZNF261 |
zinc finger protein 261 |
-2.39 |
-2.39 |
0.19 |
0.19 |
5.248917876 |
5.25675946 |
B-4:10-12 |
|
|
|
-2.41 |
-2.29 |
0.19 |
0.20 |
5.306823008 |
4.88133901 |
A-5:16-14 |
NM_000224 |
KRT18 |
keratin 18 |
-2.43 |
-2.36 |
0.19 |
0.19 |
5.404795682 |
5.14281952 |
B-7:8-16 |
NM_005780 |
LHFP |
lipoma HMGIC fusion partner |
-2.47 |
-2.68 |
0.18 |
0.16 |
5.538633731 |
6.4003583 |
C-3:5-6 |
NM_080927 |
ESDN |
endothelial and smooth muscle cell-derived neuropilin-like protein |
-2.47 |
-2.22 |
0.18 |
0.21 |
5.549855877 |
4.65363937 |
B-6:14-9 |
NM_000224 |
KRT18 |
keratin 18 |
-2.49 |
-3.07 |
0.18 |
0.12 |
5.620597234 |
8.37846833 |
D-8:5-3 |
XM_172341 |
FLJ35036 |
hypothetical protein FLJ35036 |
-2.50 |
-2.40 |
0.18 |
0.19 |
5.643607678 |
5.27784987 |
B-4:3-8 |
NM_007246 |
KLHL2 |
kelch-like 2, Mayven (Drosophila) |
-2.51 |
-2.64 |
0.18 |
0.16 |
5.696310881 |
6.23936453 |
C-5:6-16 |
NM_024074 |
MGC3169 |
hypothetical protein MGC3169 |
-2.52 |
-3.02 |
0.17 |
0.12 |
5.742923627 |
8.13173279 |
A-3:2-10 |
NM_004486 |
GOLGA2 |
golgi autoantigen, golgin subfamily a, 2 |
-2.53 |
-2.38 |
0.17 |
0.19 |
5.772987061 |
5.19065094 |
B-1:7-16 |
|
|
|
-2.59 |
-2.05 |
0.17 |
0.24 |
6.032982476 |
4.13563384 |
B-5:12-17 |
NM_005170 |
ASCL2 |
achaete-scute complex-like 2 (Drosophila) |
-2.62 |
-2.84 |
0.16 |
0.14 |
6.16397665 |
7.15336668 |
A-3:7-7 |
NM_024527 |
ABHD8 |
abhydrolase domain containing 8 |
-2.62 |
-2.27 |
0.16 |
0.21 |
6.168460111 |
4.81227412 |
A-1:12-11 |
NM_012317 |
LDOC1 |
leucine zipper, down-regulated in cancer 1 |
-2.66 |
-2.49 |
0.16 |
0.18 |
6.307614839 |
5.60637904 |
D-3:1-10 |
NM_016445 |
PLEK2 |
pleckstrin 2 |
-2.68 |
-2.58 |
0.16 |
0.17 |
6.400691717 |
5.96001472 |
B-3:17-8 |
NM_001102 |
ACTN1 |
actinin, alpha 1 |
-2.68 |
-2.50 |
0.16 |
0.18 |
6.419911166 |
5.64413645 |
D-8:14-6 |
NM_021727 |
FADS3 |
fatty acid desaturase 3 |
-2.69 |
-2.35 |
0.15 |
0.20 |
6.469132441 |
5.0921694 |
C-7:8-12 |
NM_001430 |
EPAS1 |
endothelial PAS domain protein 1 |
-2.70 |
-3.13 |
0.15 |
0.11 |
6.486214515 |
8.72684951 |
A-6:12-24 |
NM_080927 |
ESDN |
endothelial and smooth muscle cell-derived neuropilin-like protein |
-2.74 |
-2.50 |
0.15 |
0.18 |
6.691795939 |
5.67460969 |
A-4:23-22 |
|
|
|
-2.78 |
-2.18 |
0.15 |
0.22 |
6.868454225 |
4.545701 |
A-7:1-12 |
NM_003246 |
THBS1 |
thrombospondin 1 |
-2.79 |
-2.44 |
0.14 |
0.18 |
6.907748398 |
5.4118483 |
D-3:20-21 |
NM_012153 |
EHF |
ets homologous factor |
-2.83 |
-5.45 |
0.14 |
0.02 |
7.127245138 |
43.7963006 |
D-7:15-10 |
NM_012153 |
EHF |
ets homologous factor |
-2.84 |
-4.87 |
0.14 |
0.03 |
7.16989817 |
29.1565438 |
B-6:10-12 |
NM_031892 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
-2.86 |
-2.36 |
0.14 |
0.20 |
7.241210789 |
5.12084435 |
A-8:9-24 |
NM_018306 |
FLJ11036 |
hypothetical protein FLJ11036 |
-2.87 |
-2.49 |
0.14 |
0.18 |
7.305619394 |
5.63444971 |
C-8:2-5 |
NM_002318 |
LOXL2 |
lysyl oxidase-like 2 |
-2.88 |
-4.15 |
0.14 |
0.06 |
7.337897944 |
17.7454702 |
C-7:5-1 |
NM_003958 |
RNF8 |
ring finger protein (C3HC4 type) 8 |
-2.88 |
-2.33 |
0.14 |
0.20 |
7.368691284 |
5.01284359 |
D-4:18-14 |
NM_003738 |
PTCH2 |
patched homolog 2 (Drosophila) |
-2.91 |
-2.56 |
0.13 |
0.17 |
7.495722738 |
5.911008 |
B-3:1-13 |
NM_004447 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
-2.96 |
-3.50 |
0.13 |
0.09 |
7.770902194 |
11.2762852 |
B-5:10-11 |
NM_022073 |
EGLN3 |
egl nine homolog 3 (C. elegans) |
-2.96 |
-3.54 |
0.13 |
0.09 |
7.804044613 |
11.6098577 |
A-2:2-17 |
NM_138444 |
KCTD12 |
potassium channel tetramerisation domain containing 12 |
-2.97 |
-2.14 |
0.13 |
0.23 |
7.851667546 |
4.39601931 |
B-3:13-6 |
NM_007283 |
MGLL |
monoglyceride lipase |
-3.03 |
-4.63 |
0.12 |
0.04 |
8.146860637 |
24.806172 |
D-5:25-5 |
NM_005101 |
G1P2 |
interferon, alpha-inducible protein (clone IFI-15K) |
-3.06 |
-3.45 |
0.12 |
0.09 |
8.326883604 |
10.9634954 |
B-2:13-10 |
NM_005429 |
VEGFC |
vascular endothelial growth factor C |
-3.07 |
-2.51 |
0.12 |
0.18 |
8.408091946 |
5.7120402 |
A-7:17-11 |
NM_005723 |
TM4SF9 |
transmembrane 4 superfamily member 9 |
-3.09 |
-2.48 |
0.12 |
0.18 |
8.52649501 |
5.5823862 |
A-4:12-17 |
NM_153611 |
MGC20446 |
hypothetical protein MGC20446 |
-3.09 |
-3.06 |
0.12 |
0.12 |
8.539562097 |
8.34144982 |
A-8:7-11 |
NM_018192 |
MLAT4 |
myxoid liposarcoma associated protein 4 |
-3.17 |
-3.31 |
0.11 |
0.10 |
8.979387807 |
9.94098854 |
A-5:14-13 |
NM_057159 |
EDG2 |
endothelial differentiation, lysophosphatidic acid G-protein-coupled receptor, 2 |
-3.18 |
-3.02 |
0.11 |
0.12 |
9.055761789 |
8.11254773 |
D-8:14-5 |
NM_004995 |
MMP14 |
matrix metalloproteinase 14 (membrane-inserted) |
-3.19 |
-2.47 |
0.11 |
0.18 |
9.124402264 |
5.54910849 |
A-3:24-10 |
NM_001553 |
IGFBP7 |
insulin-like growth factor binding protein 7 |
-3.20 |
-2.47 |
0.11 |
0.18 |
9.194347608 |
5.52503874 |
A-5:6-16 |
NM_003803 |
MYOM1 |
myomesin 1 (skelemin) 185kDa |
-3.29 |
-3.91 |
0.10 |
0.07 |
9.774320104 |
15.0783315 |
C-2:5-5 |
NM_016233 |
PADI3 |
peptidyl arginine deiminase, type III |
-3.30 |
-4.77 |
0.10 |
0.04 |
9.862444313 |
27.3226366 |
A-8:24-7 |
NM_006517 |
SLC16A2 |
solute carrier family 16 (monocarboxylic acid transporters), member 2 (putative transporter) |
-3.37 |
-3.57 |
0.10 |
0.08 |
10.34882831 |
11.8464534 |
C-6:15-6 |
NM_012105 |
BACE2 |
beta-site APP-cleaving enzyme 2 |
-3.38 |
-2.18 |
0.10 |
0.22 |
10.43754205 |
4.54496723 |
A-6:13-6 |
NM_005672 |
PSCA |
prostate stem cell antigen |
-3.39 |
-3.50 |
0.10 |
0.09 |
10.47451419 |
11.2751577 |
A-3:8-16 |
NM_002273 |
KRT8 |
keratin 8 |
-3.45 |
-3.79 |
0.09 |
0.07 |
10.95760551 |
13.8757902 |
C-8:4-16 |
NM_007085 |
FSTL1 |
follistatin-like 1 |
-3.47 |
-6.65 |
0.09 |
0.01 |
11.11533316 |
100.251904 |
C-6:17-7 |
NM_004207 |
SLC16A3 |
solute carrier family 16 (monocarboxylic acid transporters), member 3 |
-3.51 |
-3.98 |
0.09 |
0.06 |
11.36560673 |
15.793495 |
C-4:6-11 |
NM_007203 |
PALM2 |
paralemmin 2 |
-3.51 |
-2.13 |
0.09 |
0.23 |
11.4246333 |
4.36597548 |
D-5:19-4 |
|
|
|
-3.57 |
-2.78 |
0.08 |
0.15 |
11.85742483 |
6.8516617 |
A-3:15-9 |
NM_000295 |
SERPINA1 |
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
-3.61 |
-6.57 |
0.08 |
0.01 |
12.20991141 |
95.0583386 |
A-1:1-15 |
NM_003118 |
SPARC |
secreted protein, acidic, cysteine-rich (osteonectin) |
-3.61 |
-3.10 |
0.08 |
0.12 |
12.24073208 |
8.57801881 |
A-4:15-9 |
|
|
|
-3.65 |
-2.44 |
0.08 |
0.18 |
12.56172657 |
5.44344481 |
A-3:4-14 |
NM_001654 |
TIMP1 |
tissue inhibitor of metalloproteinase 1 (erythroid potentiating activity, collagenase inhibitor) |
-3.71 |
-3.05 |
0.08 |
0.12 |
13.12721004 |
8.26900241 |
A-4:20-1 |
|
|
|
-3.95 |
-2.89 |
0.06 |
0.14 |
15.49965719 |
7.39869191 |
D-7:8-6 |
NM_021005 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
-3.98 |
-3.54 |
0.06 |
0.09 |
15.74627136 |
11.6231813 |
B-4:12-5 |
NM_001109 |
ADAM8 |
a disintegrin and metalloproteinase domain 8 |
-4.26 |
-4.02 |
0.05 |
0.06 |
19.10086542 |
16.2296299 |
D-8:17-11 |
NM_001792 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
-4.36 |
-2.91 |
0.05 |
0.13 |
20.529152 |
7.52090111 |
A-6:2-10 |
NM_182909 |
DOC1 |
downregulated in ovarian cancer 1 |
-4.39 |
-2.59 |
0.05 |
0.17 |
20.95746529 |
6.00852077 |
B-7:24-6 |
NM_024519 |
FLJ13725 |
hypothetical protein FLJ13725 |
-4.46 |
-3.29 |
0.05 |
0.10 |
21.95627756 |
9.77013135 |
D-2:10-11 |
NM_005727 |
TSPAN-1 |
tetraspan 1 |
-4.47 |
-6.29 |
0.05 |
0.01 |
22.16640231 |
78.1239771 |
D-5:8-10 |
|
|
|
-4.72 |
-4.91 |
0.04 |
0.03 |
26.31474624 |
29.984916 |
C-7:6-9 |
NM_000331 |
SAA1 |
serum amyloid A1 |
-5.15 |
-2.41 |
0.03 |
0.19 |
35.45660337 |
5.32381919 |
D-4:14-14 |
NM_012427 |
KLK5 |
kallikrein 5 |
-5.30 |
-6.55 |
0.03 |
0.01 |
39.43541733 |
93.616596 |
B-8:21-11 |
AF200348 |
D2S448 |
Melanoma associated gene |
-6.08 |
-5.15 |
0.01 |
0.03 |
67.5205415 |
35.4459189 |
D-2:7-14 |
NM_001323 |
CST6 |
cystatin E/M |
-6.35 |
-6.34 |
0.01 |
0.01 |
81.42425084 |
81.0151155 |
Table 1 L4_S0335 and L4_S0337: Common genes that are differentially regulated between Clone 11 and Clone17 in comparison with the vector transfected OSC-19. The positive logratio values are for genes that are upregulated in the clones and the negative logratio values are genes downregulated in the clones
Gene name |
Fold change |
MMP-14 |
-9.2 |
MMP-8 |
-19.5 |
KLK5 |
-93.6 |
ADAM8 |
-16.5 |
MMP3 |
+14.9 |
LEKTI |
+80.8 |
DSC2 |
+10.8 |
DSC3 |
+5.8 |
Table 2 Common genes that are differentially regulated between OSC19-LEKTI clone 11 in comparison with the OSC19-Vector clone 1. The negative fold change values are for genes that are down regulated and the positive fold change values are genes that are up regulated in the clone
Using LEKTI mAb 1C11G6, we observed that in specimens of histologically normal mucosa, LEKTI-positive staining was present in the cytoplasm of epithelial cells extending above the basal layers. Conversely, in specimens of dysplastic mucosa, LEKTI-positive staining was diminished in all layers of the epithelium. Moreover, in the majority of specimens of invasive carcinoma staining was limited to a few cells scattered within the tumor of nests of more differentiated tumor cells. Our immunohistochemical analysis of LEKTI expression in matched HNSCC patient specimens confirmed our previous findings of lost or down-regulated LEKTI mRNA transcription in similar specimens (16). We also plan to determine the expression status of severalmMP-9, mMP-14, mMP-3, and KLK5 in HNSCC tumor specimens to expand the relevance of our cell culture-based findings to patient derived tissues.
The enhancement of adhesion to ECM constituents along with the alteration in expression pattern of mMPs in LEKTI expressing clones of OSC19 demonstrates a mechanism of impaired invasive capacity. Our findings define a novel role in which LEKTI provides a critical cellular switch from stationary to migratory cell phases.
Supported in part by the NIH-NCI P50 CA097007, NIH R01 DE013954, NIH P30 CA016672, Alando J Ballantyne Distinguished Chair in Head and Neck Surgery award, Michael A. O’Bannon Endowment for Cancer Research, NIH INRS Award T32 CA060374, and AAO-HNSF Percy Memorial Grant.
The author declares no conflict of interest.

©2014 Jayakumar, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.