MOJ
eISSN: 2374-6920


Research Article Volume 6 Issue 2
1Department of Pathophysiology, Peking Union Medical College, China
2Department of Biochemistry and Molecular Biology, Beijing Normal University, China
Correspondence: Youhe Gao, Department of Pathophysiology, Peking Union Medical College, China, Tel 8610-5880-4382
Received: September 18, 2017 | Published: October 6, 2017
Citation: Wu J, Li X, Gao Y. Effects of prednisone on urinary proteome and disease biomarkers in a rat model. MOJ Proteomics Bioinform. 2017;6(2):255-261. DOI: 10.15406/mojpb.2017.06.00191
Urine is a good biomarker source for proteomics studies, but several factors may affect the urine proteome. To evaluate the possible impact of glucocorticoids drugs on the urine proteome, we investigated the effects of prednisone on urine proteome in a rat model. Urine samples were collected from control and prednisone-treated rats after drug administration. The urinary proteome was analyzed using liquid chromatography–tandem mass spectrometry (LC-MS/MS), and proteins were identified using label-free proteome quantification. A total of 523 urinary proteins were identified in rat urine. Using label-free quantification, 27 urinary proteins showed differential abundances after prednisone treatment. A total of 16 differential proteins and/or their human orthologs have been previously reported as disease biomarkers. Two differential proteins (Haptoglobin and Neutrophil collagenase) were validated via Western blot. After functional analysis, it was found that the pharmacological effects of prednisone were reflected in the urine. Prednisone treatment influences urine proteome and some disease biomarkers, and it should be taken into consideration in future disease biomarker studies.
Keywords: urine, proteomics, biomarkers, prednisone, drug treatment
Disease biomarkers are measurable changes associated with specific pathophysiological conditions that can be used to diagnose and monitor diseases, to predict prognoses and to reflect treatment efficiency. As a non-invasively accessible and relatively stable liquid, urine is a promising clinical sample source for biomarker discovery via proteomics approaches.1 Recently, urinary proteomics has received increasing attention in disease biomarker research.
Urinary proteins mainly originate from the urinary system, and the remaining proteins are plasma proteins derived from glomerular filtration.2 Currently, in addition to its extensive application in urogenital diseases, urinary proteomics have been applied to a wide range of non-urogenital diseases to exploit urinary disease biomarkers, such as cancers, cardiovascular diseases, and mental disorders, among others. However, challenges still remain in the urine biomarker discovery field. Urine is sensitive and may be affected many factors,1 such as gender,3 age,4 menstrual cycle,5 exercise,6 and smoking.7 Therefore, it is difficult to definitively determine whether potential biomarkers identified are truly related to disease.
In clinical practice, drugs can reverse disease processes and are very likely to have a significant impact on the urinary proteome and related disease biomarkers. However, due to ethical issues, it is impossible to halt drug treatment in patients during urine collection. Drugs have both therapeutic effects and side effects. Therapeutic effects cure the disease and might reduce or even eliminate disease biomarkers, whereas side effects create changes unrelated to the disease biomarker that might erroneously be considered disease biomarkers if the drug effect is not considered in a biomarker study. Knowing the impact of drugs on the urine proteome can help eliminate interference when detecting real disease biomarkers in urine.
As widely prescribed drugs, glucocorticoids are currently used to treat a wide range of diseases based on their potent anti-inflammatory, immunosuppressive and anti-neoplastic effects. One previous report indicated that approximately 0.9% of the population is using glucocorticoids at any given time.8 Among glucocorticoids, prednisone is a commonly prescribed drug that can induce an overall catabolic protein response.9 Despite the high prevalence of prednisone use in clinical practice and its strong association with protein metabolism, the effects of prednisone treatment on the urinary proteome remain unknown, which might hamper biomarker discovery for numerous diseases. In this study, an animal model was used to limit the number of confounding factors and enable observation of associated changes from fewer samples. To evaluate the possible impact of prednisone administration on the urinary proteome and related disease biomarkers, this study performed urinary proteomics analysis using LC-MS/MS in a rat model.
Experimental animals and drug administration
This study was performed on twelve male Sprague-Dawley rats (approximately 220g) obtained from the Institute of Laboratory Animal Science, Chinese Academy of Medical Science. The animals were housed in cages with six rats per cage, fed a standard laboratory diet, and provided water ad libitum. All of the rats were housed under controlled indoor temperature (221°C) and humidity (65-70%) conditions. All of the experimental protocols were reviewed and approved by Institute of Basic Medical Sciences Animal Ethics Committee, Peking Union Medical College (ID: ACUC-A02-2014-007).
After acclimatization for 3 days in cages, the rats were randomly divided into two groups. In the prednisone-treated group, rats were daily administered a prednisone solution (2mg/ml) at 4mg/kg/day via oral gavage. In the control group, rats were given a matching volume of sterile saline by intragastric administration. The rats in the prednisone group received medication every 24h for 2weeks.
Urine collection and sample preparation
Urine samples were collected from all of the rats on days 7 and 14 after the animals were individually placed in metabolic cages. The rats had free access to water but no food to avoid sample contamination during urine collection. Urine was collected within 8h of intragastric administration, and the urine volumes were measured. After collection, the urine was immediately centrifuged at 12,000g for 20min at 4°C. Urinary proteins were extracted with ethanol overnight followed by centrifugation at 12,000g for 20min. The precipitate was then resuspended in lysis buffer (8M urea, 2M thiourea, 50mM Tris and 25mM DTT). Sample aliquots were stored at -80°C for later proteomics analysis. The urine after gavage was used for subsequent laboratory biochemical analysis. The creatinine and urea nitrogen concentrations in the urine were measured at the clinical laboratory of the Peking Union Medical College Hospital.
Protein digestion
Urine samples from ten rats in two groups after gavage on day 14 were randomly selected (5 rats in each group). The urinary proteins were prepared using the filter-aided sample preparation method Wisniewski et al. Briefly, after 100µg of protein from an individual sample was denatured with 20mM dithiothreitol at 37°C for 1h and alkylated with 50mM iodoacetamide in the dark for 40min, the samples were loaded onto filter devices with a cut-off of 10kD (Pall, Port Washington, NY, USA) and centrifuged at 14,000g at 18°C. Then, after washing twice with UA (8M urea in 0.1M Tris-HCl, pH 8.5) and four times with 25mM NH4HCO3, the samples were re-dissolved in 25mM NH4HCO3 and digested with trypsin (enzyme to protein ratio of 1:50) at 37°C overnight. The peptide mixtures were desalted and dried by vacuum evaporation.
LC-MS/MS analysis
The peptide samples were re-dissolved in 0.1% formic acid to a concentration of 0.5µg/µL. For analysis, 1µg of peptide were loaded on a trap column and were separated on a reverse-phase C18 analytical column using the EASY-nLC 1200 HPLC system. The elution for the analytical column was over 120min at a flow rate of 300nL/min. Then, the peptides were analyzed with an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). MS data were acquired in high-sensitivity mode using the following parameters: data-dependent MS/MS scans per full scan with top-speed mode (3 s), MS/MS scans at a resolution of 30000 in Orbitrap, 30% HCD collision energy, charge-state screening (including precursors with a charge state of +2 to +7), dynamic exclusion (exclusion duration 30s) and a maximum injection time of 45ms. Two technical replicate analyses were performed for each individual sample.
Label-free quantification
Label-free quantitation of the proteomic data was performed using Progenesis LC-MS software (version 4.1, Nonlinear, Newcastle upon Tyne, UK). Twenty sample features (ten samples with two technical replicates) were aligned according to their retention times, and peptides with charge states of +2 to +4 were selected in the analysis. The peak lists were exported, and the data were searched against the Swiss-Prot rat database (Released in July 2014; containing 7,906 sequences) using Mascot software (version 2.5.1, Matrix Science, London, UK). The parent ion tolerance was set at 10ppm, and the fragment ion mass tolerance was set to 0.05Da. A maximum of two missed cleavage sites in the trypsin digestion was allowed. Carbamidomethylation of cysteines was set as a fixed modification, and the oxidation of methionine was considered a variable modification. For protein quantification, the total cumulative abundance of a specific protein was calculated by summing the individual abundances of unique peptides. Comparisons across different samples were performed after normalization of protein abundance using Progenesis LC-MS software.
Bioinformatics analysis and biomarker filtering
After label-free quantization, the differential proteins were screened for a fold change>1.5 and p value<0.05 between the two groups. The differentially expressed proteins were further analyzed using IPA software (Ingenuity Systems, Mountain View, CA). This analysis was used to interpret the differentially expressed proteins based on the canonical pathways, interaction networks and disease mechanisms that the proteins were expected to regulate.
The biomarker filter function in the IPA software was used to filter disease biomarkers. Additionally, we identified the human orthologs of the differentially expressed proteins using BLAST (http://www.uniprot.org/blast/). The Urinary Protein Biomarker Database is a literature curated database of protein biomarkers that have been detected in urine (Shao et al., 2011). These differentially expressed proteins and their human orthologs were searched using the Urinary Protein Biomarker Database (http://www.urimarker.com/biomarker/) to identify whether they were previously identified as candidate urinary disease biomarkers.
Western blot
Thirty micrograms of urinary protein from each sample (n=4 in each group) was loaded onto 10% SDS-PAGE gels and transferred to PVDF membranes with a transfer apparatus (Thermo Scientific, Pierce G2 Fast Blotter). After blocking in 5% milk for 1h, the membranes were incubated with primary antibodies overnight at 4°C. The primary antibodies used for validation included Haptoglobin (HPT) and Neutrophil collagenase (MMP8) (Abcam, USA). The membranes were then washed in TBST four times and incubated with secondary antibodies diluted 1:5,000 in a 5% milk solution for 1h at room temperature. The immunoreactive proteins were visualized using enhanced chemiluminescence reagents (Thermo Scientific, USA). The protein signals were scanned with an ImageQuant 400TM Imager (GE Healthcare Life Sciences, Piscataway, NJ, USA), and quantified using the AlphaEaseFC system.
Statistical analysis
Statistical analysis was performed with the Statistical Package for Social Studies (PASW statistics, SPSS, version 18.0). Data for the body weight, urine volume, and biochemical indicators in urine were presented as the mean standard deviation. All of the parameters were tested for normalization, and comparisons of these data between the control and prednisone group were performed using Student’s t-test. P-values of less than 0.05 were considered statistically significant.
Characterization of prednisone-treated rats
To investigate the effect of prednisone treatment on the urine proteome, rats were treated with prednisone or saline for fourteen days. At baseline, no significant differences of body weight were observed between two groups (222.17 7.41 vs.224.33 5.01g, prednisone vs. control). Lower body weights were observed in rats receiving prednisone therapy compared with control rats on days 7 (245.83 9.62 vs. 255.50 7.04g) and 14 (279.50 10.84 vs. 292.33 9.27g). However, this difference was not statistically significant (Figure 1a).
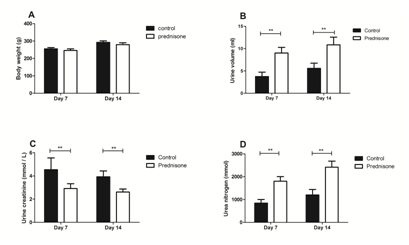
Figure 1 Body weights and urinary biochemical indicators in prednisone-treated and control rats
Urine samples were collected within 8h of intragastric administration in the two groups, and urine volumes and biochemical indicators were measured. As shown in Figure 1b, rat urinary volumes increased significantly after prednisone treatment compared with controls (9.02 1.25 vs. 3.75 0.96ml, on day 7; 10.85 1.70 vs. 5.58 1.67ml, on day 14, both p-values<0.01). Meanwhile, the urine creatinine concentration was decreased in the prednisone group (Figure 1c, p<0.05). As shown in Figure 1d, the excretion of urea nitrogen was significantly increased in urine from the prednisone group (1800.69 203.88 vs.843.21 156.86mMol, on day 7; 2,417.41 265.41 vs. 1,204.81 233.58mMol, on day 14, both p-values<0.01).
Changes to the urine proteome after prednisone administration
For proteomics analysis, ten urine samples were randomly selected from the prednisone and control groups on day 14. Using the label-free quantification by the Progenesis LC-MS software, 523 urinary proteins were identified with a 1% false discovery rate (FDR) at the protein level. The criteria for identifying differentially expressed proteins included a fold change>1.5 and p value<0.05 between the two groups. Ultimately, 27 differentially expressed proteins were identified in urine with at least 2 unique peptides used for quantitation. Specifically, 12 proteins showed increased relative abundance, and 15 proteins showed decreased abundance (Table 1).
Uniprot |
FC |
Description |
P |
Human Ortholog |
IPA |
Urinary Protein Biomarker Databaseb |
|
|---|---|---|---|---|---|---|---|
Biomarker-animal |
Biomarker-human |
||||||
P02764 |
1.77 |
Alpha-1-acid glycoprotein |
0.004 |
P02763 |
Yes |
↑in DN |
↑in T1DM, DN, renal AR, AKI, acute appendicitis, vasculitis and pre-eclampsia |
P05545 |
1.61 |
Serine protease inhibitor A3K |
0.009 |
P01011 |
- |
↓in acute renal failure and nephrotic syndrome |
↑in T2DM, renal AR, non-small cell lung carcinoma and acute appendicitis |
P09006 |
2.42 |
Serine protease inhibitor A3N |
0.001 |
P01011 |
Yes |
- |
↑in T2DM, renal AR, non-small cell lung carcinoma and acute appendicitis |
P02780 |
2.66 |
Secretoglobin family 2A member 2 |
0.053 |
- |
- |
↓in nephrotic syndrome |
- |
P06866 |
2.08 |
Haptoglobin |
0.047 |
P00738 |
Yes |
↑in nephrotic syndrome |
↑bladder transitional cell carcinoma, AKI, DN, and T2DM |
P36374 |
2.01 |
Prostatic glandular kallikrein-6 |
0.047 |
- |
- |
↓in acute renal failure |
- |
Q9EQV6 |
1.90 |
Tripeptidyl-peptidase 1 |
0.001 |
O14773 |
- |
- |
- |
O88766 |
1.69 |
Neutrophil collagenase (MMP8) |
0.005 |
P22894 |
Yes |
- |
- |
P02782 |
2.79 |
Prostatic steroid-binding protein C1 |
0.038 |
- |
- |
- |
- |
O54715 |
1.85 |
V-type proton ATPase subunit S1 |
0.001 |
Q15904 |
- |
- |
- |
Q9R0J8 |
1.51 |
Legumain |
0.045 |
Q99538 |
- |
- |
- |
P84039 |
2.11 |
Ectonucleotide pyrophosphatase/phosphodiesterase family member 5 |
0.010 |
Q9UJA9 |
- |
- |
- |
P23680 |
-1.68 |
Serum amyloid P-component |
0.006 |
P02743 |
Yes |
↑in anti-GBM nephritis and lupus nephritis |
↑in intestinal mucosal injury |
P04764 |
-1.57 |
Alpha-enolase |
0.011 |
P06733 |
Yes |
- |
↓in Dents disease |
P81828 |
-1.61 |
Urinary protein 2 |
0.039 |
- |
- |
↓in nephrotic syndrome |
- |
P81827 |
-1.68 |
Urinary protein 1 |
0.038 |
- |
- |
↓in nephrotic syndrome and glomerulonephritis |
- |
P35444 |
-1.62 |
Cartilage oligomeric matrix protein |
0.011 |
P49747 |
Yes |
↑in osteoarthritic |
- |
P01830 |
-1.90 |
Thy-1 membrane glycoprotein |
0.036 |
P04216 |
Yes |
- |
↑in prostate cancer |
O35112 |
-1.51 |
CD166 antigen |
0.004 |
Q13740 |
Yes |
- |
↑in T1DM |
P52590 |
-1.99 |
Nuclear pore complex protein Nup107 |
0.007 |
P57740 |
- |
- |
↓in IgA nephropathy |
P15473 |
-1.53 |
Insulin-like growth factor-binding protein 3 |
0.017 |
P17936 |
Yes |
- |
↑prostate cancer |
P11030 |
-1.56 |
Acyl-CoA-binding protein |
0.029 |
P07108 |
- |
- |
↑in Dents disease |
P97603 |
-1.50 |
Neogenin (Fragment) |
0.009 |
Q92859 |
- |
- |
- |
P80202 |
-1.89 |
Activin receptor type-1B |
0.046 |
P36896 |
- |
- |
- |
P08649 |
-1.66 |
Complement C4 |
0.019 |
P0C0L4 |
- |
- |
- |
P16310 |
-1.58 |
Growth hormone receptor |
0.039 |
P10912 |
- |
- |
- |
Q5FVR0 |
-1.51 |
T-cell immunoglobulin and mucin domain-containing protein 2 |
0.020 |
- |
- |
- |
- |
P14046 |
-1.71 |
Alpha-1-inhibitor 3 |
0.003 |
P01023 |
- |
- |
- |
Q01205 |
-1.61 |
2-oxoglutarate dehydrogenase complex component E2 |
0.026 |
P36957 |
- |
- |
- |
Table 1 Details of the identified urinary proteins altered by prednisone treatment
aDisease biomarkers filtered using IPA. bUrinary disease biomarkers from the Urinary Protein Biomarker Database identified in animal models and human diseases.
Abbreviations: T1DM: Type 1 Diabetes; T2DM: Type 2 Diabetes; DN: Diabetic Nephropathy; AKI: Acute Kidney Injury; AR: Allograft Rejection.
To identify major diseases and bio-functions of differential proteins involved, Ingenuity Pathway Analysis (IPA) Software was used for canonical function enrichment analysis. As shown in Figure 2, the principal diseases affected by the differential proteins included the inflammatory response, metabolic disease and cancer. The principal bio-functions of the differential proteins included cell death and survival, protein metabolism (degradation and synthesis), lipid metabolism, amino acid metabolism and carbohydrate metabolism.
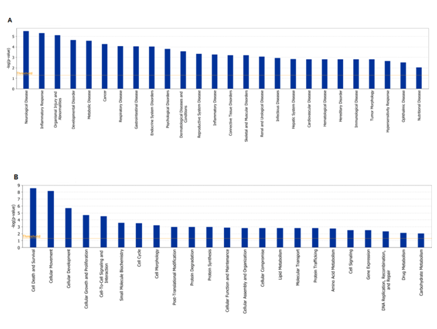
Figure 2 Analysis of principal diseases and bio-functions in which the differentially expressed proteins participate using IPA.
Significance values were calculated based on Fisher’s right tailed exact test, and the –log(p-value) was calculated and is displayed on the y-axis of the bar chart. The taller the bar, the more significant the pathway effect.
Two up-regulated proteins (Haptoglobin and Neutrophil collagenase) identified in our proteomic study with commercially available antibodies were selected to be validated by semi-quantitative western blot analysis (Figure 3). The changes of both proteins in Western Blot experiment were consistent with the LC-MS/MS results.
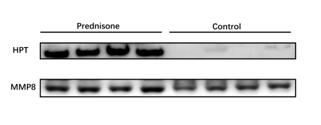
Figure 3 Western blot analysis of HPT and MMP8 in the urine samples after prednisone administration.
A total of eight samples were used for validation (n = 4 in each group). The intensity of each protein before and after prednisone administration was analyzed by Mann-Whitney test.
Effects of prednisone treatment on relevant disease biomarkers in urine
Using the biomarker filtering tool in IPA, 10 differential proteins were identified as disease biomarkers. Because orthologs have similar bio-functions across different species, we converted the rat proteins to human proteins, and 23 differential proteins had human orthologs. When the selected differential proteins and their human orthologs were searched against the Urinary Protein Biomarkers Database, 16 proteins were annotated as candidate urine biomarkers in previous studies. The expression trends of these biomarkers in various diseases are shown in Table 1. The expression of 8 differential proteins were completely reversed after prednisone treatment compared with their expression in specific pathological conditions, including Secretoglobin family 2A member 2, Prostatic glandular kallikrein-6, Serum amyloid P-component, Cartilage oligomeric matrix protein, Thy-1 membrane glycoprotein, CD166 antigen, Insulin-like growth factor-binding protein 3, and Acyl-CoA-binding protein. Meanwhile, the expression of 7 differential proteins in our study after prednisone treatment were similar to levels in specific disease conditions, including Alpha-1-acid glycoprotein, Serine protease inhibitor A3N, Haptoglobin, Alpha-enolase, Urinary protein 1, Urinary protein 2, and Nuclear pore complex protein Nup107.
In this study, we evaluated the possible impact of prednisone treatment on the urinary proteome and related disease biomarkers in a rat model. The doses of prednisone used in this study were according to a previous study to mimic clinical practices.10 Changes of body weights and urine biochemical indicators in prednisone-treated rats were consistent with pharmacological effects of prednisone. Reduced body weight might be due to growth inhibition caused by glucocorticoid treatment.11 The increased urine volume and decreased creatinine concentration after prednisone treatment were induced by the diuretic effect of glucocorticoids. Additional, prednisone treatment induces an overall catabolic protein response and accelerate protein breakdown,9 which can lead the body to dispose of excess nitrogen waste as urea in urine.
The strong association between prednisone treatment and protein metabolism may also contribute to the changes in the urinary proteome. Several differential proteins identified in current study were reported to be regulated by glucocorticoids in previous studies, such as the serine protease inhibitor A3N,12 alpha-1-acid glycoprotein,13mMP8,14 growth hormone receptor15 insulin-like growth factor-binding protein 316 and acyl-CoA-binding protein,17 suggesting the reliability of the proteomics results. Moreover, two differentially expressed proteins (Haptoglobin and Neutrophil collagenase) in our study were further validated by Western Blot. The results suggested these urinary proteins were glucocorticoid responsive, similar to thrombospondin-1,18 which has potential clinical utility as a glucocorticoid activity biomarker in urine.
Prednisone is widely used in clinical practice based on its potent anti-inflammatory, immunosuppressive and anti-cancer effects. The enriched bio-function of differential proteins was consistent with the regulatory actions of prednisone in inflammation, immunity, metabolism and cancer, suggesting that the pharmacological effects of prednisone were reflected in the urine proteome after prednisone administration. As a good biomarker source, urine has the potential to reveal many types of changes (physiological, pathological, and pharmacological changes) in the body. Our results suggested the application of urinary proteomics may assist in identification of non-invasive biomarkers to detect drug efficacy in future studies.
One important issue in the use of urine to detect biomarkers is that many factors can affect the urine proteome. Previous studies showed that several common physiological factors influence the urine proteome.1 Compared with physiological factors, pharmacological agents are more important factors that should also be taken into consideration. In clinical practice, drugs can reverse the disease process and are very likely to have a significant impact on the urinary proteome and related disease biomarkers. Of note, physiological factors can often be well-matched for biomarker research. However, due to ethical issues, it is impossible to halt drug treatment in patients during urine collection or to give healthy volunteers drugs they do not need. Knowing the impact of drugs on the urine proteome can help us eliminate interference when detecting real disease biomarkers in urine. In previous studies, anticoagulants19 and the α1-adrenergic receptor antagonist20 were reported to influence the urine proteome and related biomarkers. Likewise, in the current study, 27 proteins were differentially expressed after prednisone treatment, and 16 of these proteins were reported as disease biomarkers, suggesting the potential impact of prednisone treatment on urine biomarker identification via proteomics approaches.
Notably, the expression of some candidate biomarkers were completely reversed after prednisone treatment compared with their expression in specific pathological conditions (Table 1). These proteins might serve as candidate therapeutic biomarkers for monitoring treatment effectiveness or reflecting glucocorticoid sensitivity. For example, up-regulated serum amyloid P (SAP) in the urine was identified as a biomarker of anti-glomerular basement membrane disease and lupus nephritism,21 and increased cartilage oligomeric matrix protein (COMP) in urine was observed in osteoarthritis.22 Both proteins were downregulated in urine after prednisone treatment in our study; thus, these proteins might indicate the effectiveness of glucocorticoids in treating these diseases. Thus, previous studies with patients receiving drug treatments might have underestimated or not identified changes to candidate disease biomarkers. By contrast, the expression of several candidate biomarkers in our study after prednisone treatment was similar to levels in specific disease conditions. These changes might not be relevant to the treatment effects of prednisone for these diseases and might even be erroneously considered disease biomarkers if the drug effect was not considered. Therefore, the effects of prednisone treatment should be taken into consideration in future disease biomarker studies.
In summary, prednisone is a widely prescribed drug in clinical practice, and its effects are strongly associated with protein metabolism. In this study, the significant changes to protein abundance identified by mass spectrometry suggested a large impact of prednisone treatment on urine proteome and related urinary biomarkers. Therefore, changes to the urine proteome and disease biomarkers from prednisone treatment should be considered in future biomarker studies.
This work was supported by the National Key Research and Development Program of China (2016YFC1306300); the National Basic Research Program of China (2013CB530805); Beijing Normal University (11100704); and Beijing Natural Science Foundation (Nos. 7173264, 7172076). The raw mass spectrometry data are available in the Figshare repository (https://doi.org/10.6084/m9.figshare.3486035.v1).
The authors declare that there is no conflict of interests regarding the publication of this paper.

©2017 Wu, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.