MOJ
eISSN: 2374-6920


Research Article Volume 3 Issue 6
School of Sciences, RMIT University, Australia
Correspondence: Ian Macreadie, School of Science, RMIT University, Bundoora; Victoria 3083, Australia, Tel +613 9925 6627, Fax +61 3 9925 7110
Received: June 19, 2016 | Published: July 15, 2016
Citation: Dorji T, Bosser S, Siriwardena VS, et al. Can yeast salvage folate? bioinformatics suggests yes! MOJ Proteomics Bioinform. 2016;3(6):140-147. DOI: 10.15406/mojpb.2016.03.00104
Folate is an important nutrient for cell growth and division in all organisms. Many microorganisms including prokaryotes and eukaryotes can synthesize their own folate while humans cannot. To assimilate folate into the cell a folate transporter is required for uptake through the cell membrane. Three types of folate transporters are known to exist in humans to facilitate the uptake of folate: Reduced Folate Carrier (RFC), Proton-Coupled Folate Transporter (PCFT) and Folate Receptors (FR). Although yeast cells can synthesize their own folate, laboratory evidence suggests that it can also utilize folate from exogenous source. However, little is known about the presence of folate transporters and other proteins involved in utilization of folate in yeasts. Hence using various bioinformatics analysis tools we attempted to determine the types of folate transporter present using genome sequences of well-studied yeasts from Entrez Database, Yeast Genome Database and Candia Genome Databases. Bioinformatics suggests the folate transporter present the yeast is similar to human PCFT with significant identity. All yeast possessed genes to encode the putative proteins which are similar to human PCFT. Saccharomyces cerevisiae’s putative protein had the highest identity match (identity=30.2%, E-value≤0.05) while Candida glabrata had the lowest (identity=22.6%, E-value:<0.05). Phylogenetic analysis showed yeast PCFT being distant from human PCFT but the conserved domain and hydropathy analysis indicated that it performs similar function to the human PCFT. Although bioinformatics suggests the presence of PCFT in yeast, genetic studies in the future could further help to understand the PCFT. If laboratory results are consistent with our finding the yeast could be used as a model organism to study many diseases in humans related to PCFT.
Keywords: folate transporter, FGCP, PCFT, yeast, conserved domain, phylogenetic analysis, bioinformatics, heme carrier protein, folate receptor-alpha, deoxyribonucleic acid, proton-coupled folate transporter, thymidylate synthase, monoglutamate, candida albicans,computational analysis, genomic analyses, genome, proteins, escherichia coli, caenorhabditis elegans
BLAST, basic local alignment search tool; CD, conserved domain; DHFR, dihydrofolate reductase; DNA, deoxyribonucleic acid; FGCP, folylpoly-gamma-glutamate carboxypeptidase; HCP, heme carrier protein; FR-α, folate receptor-alpha; FR-β, folate receptor-β; GCP, glutamate carboxypeptidase; MFS, major facilitator superfamily; MTHF, methyl tetrahydrofolate; NCBI, national center for biotechnology information; PCFT, proton-coupled folate transporter; RCF, reduced folate carrier; T-COFFEE, tree-based consistency objective function for alignment evaluation; THF, tetrahydrofolate; TS, thymidylate synthase
Folate (also known as vitamin B9 Folate) is a water soluble micronutrient which is essential for all living organisms. It plays an important role in cell division and is required for nucleic acid synthesis, methylation of DNA, and other metabolic regulations. Folate deficiency is associated with many chronic diseases including cancers and developmental disorders.1,2 Although probiotic bacteria such as Lactobacilli and Bifodobacterium, plants and yeasts are known to produce folate, many other living organisms, including human cells, have no ability to produce folate on their own and rely on exogenous sources from the diet.3,4 Several well-studied yeasts species such as Pichia, Saccharomyces, Candida, Klyveromyces, Cryptococcus, Pseudozyma and Rhodotorula are known to produce folate. Candida glabrata, for instance, was observed to produce high amount of folate within few days of fermentation. Folate production was observed to increase by several folds when the organism was fermented in togwa, an indigenous maize-based porridge of Tanzania.4,5
Folate uptake and transport into the cell cytoplasm involves several biological processes. The exogenous folate sources mostly exist in polyglutamate form which needs to be hydrolyzed into monoglutamates in order to be transported into the cells through the cell membrane. The folypoly-ɣ-glutamate carboxypeptidase (FGCP), which is encoded by glutamate carboxypeptidase II genes (GCP2), is responsible for the hydrolysis of polyglutamate folate into monoglutamate, primarily 5-methyltetrahydrofolate monoglutamate (5-MTHF). Once hydrolyzed, it is transported into the cells by a high-affinity Proton Coupled Folate Transporter (PCFT1), also called Heme Carrier Protein I (HCP1) and folate receptors composed of alpha (FR-α) and Beta (FR-β) subunits which have high affinity to monoglutamates.6 Also, other important folate carrier proteins such as the Reduced Folate Carrier (RCF) are considered to be a major folate transporter system and play important role in treatment of different diseases in humans.7 Once MTHF monoglutamate is inside the cell cytoplasm it donates a carbon to convert dUMP to dTMP by thymidylate synthase (TS) which results in conversion of MTHF to dihydrofolate (DHF). The DHF is reduced to tetrahydrofolate (THF) by dihydrofolate reductase (DFHR).8
Although yeasts are known to produce their own folate they were also found to possess the capability to utilize folate from exogenous source. When the genes which encodes for folate are knocked out and grown on medium containing folic or folinic acid (an alternative form of folic acid) it was observed that these mutant yeast strains could grow and form multiple buds.9 Computational analysis of the Major Facilitator Superfamily (MFS) of Candida albicans detected the presence of PCFT/HCP family represented by single member or f19.6976 which was homologous to PCFT in human.10 These observations indicate that yeasts can salvage folate from external sources and utilize it in the absence of endogenous folate synthesis using folate transporters. However, the presence of folate transporters and its mechanism of uptake of folate by yeast cells are rarely studied and currently very limited information is available about the presence of folate transporters in yeasts. Therefore, we attempted to investigate the presence and type of folate transporter through genomic analyses of yeast species using various bioinformatics analytical tools. If the evidence of presence of the folate transporter is established, this study would enable further laboratory research on the folate transport system to have a deeper understanding on the mechanism of folate transport in yeast cells. The information derived from the genomic and laboratory analysis may be useful as a model organism to study folate related diseases in humans.
A series of computational techniques were used in present study. We commenced by investigating various genes encoding for hydrolase enzymes as the hydrolases play important role in converting polyglutamate to monoglutamate in order to transport folate across the membrane in Entrez (http://www.ncbi.nlm.nih.gov). Then gene loci encoding for different types of folate transporters were identified in humans and other well studied organisms using Entrez database. These well-known folate transporter proteins were used as queries to perform series of BLAST searches in the Candida Genome Database (www.candidagenome.org) and Saccharomyces Genome Database (www.yeastgenome.org) to identify similar proteins in yeast. Once the putative folate transporter was identified, further BLAST searches were conducted to determine presence of homologous proteins in other organisms such as Escherichia coli and Caenorhabditis elegans. Then, PSI BLAST was performed between high scoring homologous proteins to determine the degree of homology among yeast and to find distantly related proteins not detected by BLASTp searches. Homologous proteins were selected for multiple sequence analysis (MSA) using Clustal Omega and T-COFFEE (http://www.ebi.ac.uk/). The ORFs identified were used for domain analysis to determine the conserved domain (CD) regions. Domain analysis was first conducted among different yeast strains to determine the conserved region in the amino acid sequence encoding for the transporter protein. Additional domain analysis was conducted with the sequences from yeasts and other organisms mentioned above to see conserved regions between yeast and other organisms. Phylogenetic analyses were conducted among yeasts and a few eukaryotic organisms including human based on MSA results determine the evolutionary relationships for these organism with a shared common ancestor. The Kyte-Doolittle method (http://www.tcdb.org/progs/?tool) was used to determine hydrophobicity and Protter software (http://wlab.ethz.ch/protter) was used to study topology of transmembrane region of these proteins in yeasts.
Folate is an important micronutrient for all living organisms. Exogenous folate is transported into cells by folate transporters. We conducted various computational analyses using various bioinformatics techniques and software programs to investigate for folate transporter in yeast cells. A BLAST search using a known hydrolase sequence revealed that the yeasthas a folylpoly-ɣ-glutamate carboxypeptidase (FGCP) with similarity to the human GCP 2 (NP_004467.1) enzyme. MSA performed using Candida albicans, Candida glabrata and Saccharomyces cerevisiae showed these yeasts have identity of 31.73%, 24.56% and 26.47% respectively to human GCP2 (Figure 1).
A query search for folate receptors and the reduced folate carrier did not detect any match when BLASTp search was performed in the Saccharomyces and Candida genome databases. However, a significant match was observed when the human proton-coupled folate transporter (hPCFT)/ heme carrier protein (HCP) (NP_542400.2 ) when a protein query was BLAST searched in the Candida Genome and Saccharomyces Genome Databases. Blast search revealed that the hPCFT has homologues in most of the well-studied yeasts. This study analyzed the protein homologues in Candida albicans, Candida glabrata, Saccharomyces cerevisiae, Kluyveromyces lactis, Penicillium rubens Wisconsin and Aspergillus niger. The hPCFT protein sequence was homologous to uncharacterized membrane protein (EJS43219.1) of Saccharomyces cerevisiae (identity=30.2%, E-value= <0.05), predicted MFS membrane transporter member of PCFT/HCP family (CDGID: CAL0000192135) of Candida albicans (orf19.6976) (identity=25%, E-value: <0.05), hypothetical protein (XP_451852.1)of Kluyveromyces lactis (identity=25%, E-value: <0.05), putative protein (CGDID:CAL0137415) of Candida glabrata (identity=22.6%, E-value: <0.05), putative protein (XP_002560362.1) of Penicillium rubens Wisconsin (identity=25%, E-value: <0.05) and MFS transporter (XP_001397281) of Aspergillus niger. Conserved domain analysis of this ORF detected 15 domains and all the domains from different yeasts and A. niger belong to the major facilitator superfamily (MFS) (cl21472) (Figure 2).
MSA using Clustal Omega 2.1 and T-COFFEE revealed highly conserved amino acids residues in the sequences of PCFT proteins among yeasts. G, D, R, T, Q, L, S, A, S, Y, E, P, W and K were highly conserved in these sequences (Figure 3). Further MSA was conducted with the yeast putative proteins, hPCFT, and putative proteins in E. coli and C. elegans. We observed that G, D, R, E, W and Y residues were conserved among these species (Figure 4). The homologous amino acids sequences representing PCFT protein in yeasts, human, nematodes and bacteria were used to generate a phylogenetic tree to understand similarities among yeasts, humans, bacteria and nematodes (Figure 5). E. coli (WP_061811798) and C. elegans (NP_500781.2)were chosen to represent bacteria and nematodes. Phylogenetic analysis indicated that the PCFT proteins of humans, C. elegans and E. coli are relatively more dissimilar to those from yeast species. Among the yeast species C. glabrata is more similar to S. cerevisiae than K. lactis while C. albicans, P. rubens Wisconsin and A. niger appear to be less similar to S. cerevisiae. P. rubens Wisconsinand A. niger form a clade that is a sister group to a clade consisting of C. albicans, K. lactis, C. glabrataand S. cerevisiae. C. albicans appears to have obtained genes encoding the protein from K. lactis, C. glabrata and S. cerevisiae through horizontal gene transfer.
Transmembrane proteins are the membrane proteins that are found in cell membrane to facilitate transport of specific substance into the cytoplasm of a cell. Figure 6 shows the hydropathy index and transmembrane of PCFT protein of in the yeast cells. The sliding window analysis of protein sequence using the Kyte-Doolittle method was used to determine hydrophobic and hydrophilic regions. Each amino acid residue had a variable hydropathic index varying from negative (hydrophilic) to positive (hydrophobic). However, the predicted transmembrane protein was a mostly hydrophobic domain. Earlier studies had also determined that membrane associated folate binding protein are extremely hydrophobic.11 This could be due to the fact that the PCFT protein sequence contains amino acids with hydrophobic functional group such as alanine, glycine, serine and tyrosine. Topology analysis of the transmembrane protein from yeasts also revealed 12 membrane spanners with a long N-terminal and short C-terminal made of about 79 and 8 amino acids respectively (Figure 7A). In comparison, the human PCFT has a shorter N-terminal segment with a similar C-terminal segment length (Figure 7B). It also appears that both N-terminal and C-terminal segments are located inside the cytosolic region. Similarly immunohistochemical analysis of the human and rodent PCFTs indicated that both the C-terminal and N-terminal regions are located inside the cytoplasm.12
BLAST search and homology study suggests that the PCFT is the predominant folate transporter present in yeasts and that FGCP hydrolyzes folate prior to transport by PCFT. FGCP is a membrane bound metalloenzyme that catalyzes the breakdown of polyglutamated folate to monoglutamated folate in order to enable cells to expedite transport of folate into the cytoplasm. Structurally this class II membrane protein is located in periphery of cell membrane and has single membrane spanner made up of about 20 amino acids and short amino terminus.13,14 PCFT, also known as HCP was previously thought to function in heme transport activity but later found to function in transport of proton-dependent folate transport.15 It was found to be a primary folate transporter and its absence resulted in reduced folate uptake in an in vivo experiment of mice.16 Mutation in PCFT in humans is associated with many chronic and hereditary diseases. Hereditary folate malabsorption, neural tube defects, hyperhomocysteinemia and oral cleft are some of the defects due to mutation in hPCFT protein.17-19 The identity percentage of PCFT in yeasts and humans is quite low but significant. This could be because they are quite dissimilar organisms, as also indicated by the phylogenetic analysis. However conserved domain analysis, clearly indicates that these proteins perform similar functions. PCFT belongs to largest membrane protein super family, MFS which are involved in symport and uniport of different substances across the cell membrane into the cytoplasm including sugars, drugs, peptides, etc.20 The analysis of transmembrane proteins of yeast also suggested similarity to human PCFT with equal numbers of membrane spanners and both the sequences showing similarity in hydropathy characteristics. These data suggest that PCFT is present in the membrane of yeast cells which is facilitating the transport of folate from exogenous sources. The results obtained by laboratory experiments that determined the ability of yeast cells to utilize exogenous folate when their genes encoding for folate synthesis are knocked out also supports our findings.21-24
Although bioinformatics suggests yeast can salvage folate from exogenous source, laboratory studies carried out by deleting the genes encoding PCFT could provide crucial evidence in support of PCFT being the main folate transporter in the yeast cells. Such a study would also provide an opportunity to look for the presence of other folate transporters as well as folate receptors and the reduced folate carrier in yeast.

Figure 1 Percent Identity Matrix showing human Glutamate carboxypeptidase 2 homologues in Candida albicans, Candida glabrata and Saccharomyces cerevisiae. Percent Identity Matrix generated using Clustal 2.1
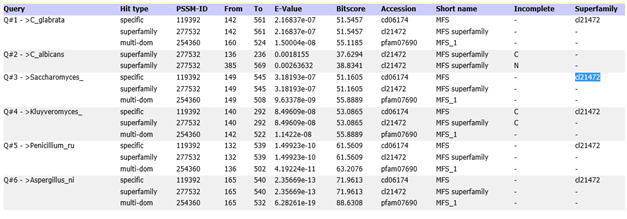
Figure 2 Conserved domain analysis of putative protein sequence in yeasts showing significant identity match to MFS protein family.



Figure 3 MSA showing conserved amino acids in PCFT of C. glabrata, C. albicans, Saccharomyces cerevisiae, Kluyveromyces lactis, Penicillium rubens Wisconsin and Aspergillus niger.




Figure 4 MSA showing conserved amino acids in PCFT of C. glabrata, C. albicans, Saccharomyces cerevisiae, Kluyveromyces lactis, Penicillium rubens Wisconsin and Aspergillus niger, E. coli and C. elegans.

Figure 5 Phylogentic Analysis showing evolutionary relationship among yeasts, human and E. coli and C. elegans.
In conclusion, folate is an important micronutrient for all yeast species. Yeasts can produce its own folate; however, it can also utilize folate from exogenous source. To salvage folate, yeasts require a folate transporter. Bioinformatics analysis strongly suggests that yeasts can salvage folate from exogenous source using a hydrolase and a PCFT-like putative protein which has significant identity to PCFT in human. Future studies using yeast could provide additional evidence to support our findings. If the existence of PCFT in yeast is further proven by laboratory study, yeast could be deployed as a model organism to study many chronic and hereditary diseases related to PCFT protein.
None.
The author declares no conflict of interest.

©2016 Dorji. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.