MOJ
eISSN: 2374-6920


Research Article Volume 2 Issue 2
1Department of Pharmacology and Toxicology, University of Alabama at Birmingham, USA
2Division of Adolescent Medicine, Children's Hospital Medical Center, USA
3Department of Environmental Health, University of Cincinnati College of Medicine, USA
4Division of Preventive Medicine, UAB
5UAB Comprehensive Cancer Center, University of Alabama at Birmingham, USA
6The Irma Russo Breast Cancer Research Laboratory, Fox Chase Cancer Center-Temple University Health System, USA
Correspondence: Coral A Lamartiniere, Department of Pharmacology and Toxicology, University of Alabama at Birmingham, 1670 University Boulevard, VH 241, Alabama 35294, USA, Tel 2057900573, Fax 2059348240
Received: December 30, 2014 | Published: March 10, 2015
Citation: Wang J, Betancourt A, Jenkins S, et al. Altered blood proteome in girls with high urine concentrations of bisphenol a, genistein, mono-ethyl hexylphthalate and mono-benzyl phthalate. MOJ Proteomics Bioinform. 2015;2(2):44-57. DOI: 10.15406/mojpb.2015.02.00040
Children exposed to endocrine disruptors are hypothesized to be susceptible for cancer development later in life. Identifying functional biomarkers of specific exposures may indicate predisposition for this disease. The objectives of this study were to identify protein biomarkers of 1) effect and 2) susceptibility for cancer from the blood of girls exposed to select environmental chemicals. In prepubertal girls, urine concentrations of bisphenol A (BPA), genistein, mono-ethyl hexylphthalate (MEHP) and mono-benzyl phthalate (MBzP) were used to identify girls in the top quintile of exposure for each of these environmental chemicals, and age-matched prepubertal girls with urine analyte concentrations below the median. Blood samples of these girls were depleted of the seven most abundant proteins using human-specific affinity spin columns. Using isobaric Tandem Mass Tags and quantitative mass spectrometry (TMT-MS), 51, 34, 57 and 47 differentially expressed proteins were identified from the blood of prepubertal girls with high urine concentrations of BPA, genistein, MEHP and MBzP, respectively, compared to controls. The data demonstrates the potential of proteomic technology to not only provide biomarkers of effect from aminimally invasive source of biological material, blood, but to identify protein molecules that are intimately involved in the pathobiology of cancer. The differentially regulated cancer associated proteins in girls with high concentrations of BPA and genistein are consistent with reported roles of BPA in carcinogenesis and of genistein in mammary cancer prevention, respectively.
Keywords: girls, blood, biomarkers, proteomics, toxicology, cancer
AIN, american institute of nutrition; BBP, butyl benzyl phthalate; BPA, bisphenol a; BW, body weight; DEHP, di-(2-ethylhexyl)phthalate; DLC1, deleted in liver cancer 1; DNM3B, DNA (cytosine-5)-methyltransferase 3b; ECE-1, endothelium-converting enzyme;EIF3, eukaryotic initiation factor 3; FDR, false discovery rate; KLF10, krueppel-like factor 10; IGFBP-3, insulin growth factor binding protein 3; Mbzp, mono-benzyl phthalate; MEHP, mono-ethyl hexylphthalate; MKK4, mitogen-activated kinase kinase 4; MS, mass spectrometry; Mudpit, multidimensional protein identification technology; NMU, n-nitroso-n methylurea; PANTHER, protein analysis through evolutionary relationships system; PRDM5, pr domain zinc finger 5; PQD, pulsed q dissociation; RAD54B, rad54 homolog b; RB1, retinoblastoma-associated protein; SAM, significance analysis of microarray; TMT, tandem mass tags; TRPC5, transient receptor potential channel 5
Natural and man-made chemicals may act as endocrine disruptors, interacting with hormone receptors, influencing synthesis and metabolism of hormones and transcription factors, and affecting the hypothalamic-pituitary-gonadal axis1 One example is BPA, which is used in the manufacture of children’s polycarbonate milk bottles, toys, lining of food and soft drink cans, cash register receipts and many plastic products. BPA can leach from these containers and products and be ingested.2 Exposure of the American population to BPA has been detected in 92.6% sampled urine, with concentrations of 0.4 to 149ng/mL.3,4 Epidemiological studies have shown an association between BPA exposure and cardiovascular disease,5 obesity,6 diabetes,7 and cancer.8–10 Durando et al.,11 have shown that prenatal exposure to BPA coupled to a sub-carcinogenic dose of N-nitroso-N methylurea (NMU) resulted in an increased percentage of pre neoplastic and neoplastic lesions in the rat mammary gland. Murray et al.,12 showed that fetal exposure of rats to BPA induced mammary gland ductal hyperplasia and carcinoma in situ. Jenkins et al.,13 reported that prepubertal exposure to BPA increased susceptibility for chemically-induced mammary cancer in rats.
Phthalates are used in the manufacture of polyvinylchloride water pipes, vinyl and carpet tiles, artificial leather, detergents, lubricating oils, certain adhesives, plastic food wrap, children’s toys and found in cosmetics and shampoos.14 In the United States, it has been estimated that the levels of phthalates in foods can vary between 50–500mg/kg.15,16 The best estimate of exposure to the general public is 2μg/kg body weight (BW)/day from food in adults, with exposures to infants and children up to 3-fold higher.17 A significant delay in the age at vaginal opening (approximately 2days) in rat offspring from dams exposed to 15mg di-(2-ethylhexyl)phthalate (DEHP)/kg bw/day during pregnancy and during lactation has been reported.18 are some of the adverse effects seen in adult female rats exposed to high doses of DEHP.19 Using a recombinant yeast screen for estrogenic activity, butyl benzyl phthalate (BBP) was found to be the most potent phthalate, although approximately 1x106 less than 17beta-estradiol.20 Phthalates that were estrogenic in the yeast screen are also mitogenicin human breast cancer cells.21 A study of girls with early the larche suggests a possible association between plasticizers with known estrogenic and anti androgenic activity and the cause of premature breast development in the human female population.22 Certain phthalates have been reported to enhance liver and skin carcinogenesis in rodents, and to act as weak estrogens toward human breast cancer cells.22–24
Genistein is an isoflavone component of soy. Soy-based diets are high in phytochemicals and quantitative results indicate that phytoestrogens are normal constituents of human urine from participants consuming large amounts of soy products (tofu, soy flour, soy milk, tempeh, etc).25 Asian women, consuming a diet high in soy products, have a low incidence of breast cancer.26,27 Yet, Asians who immigrate to the United States and adopt a western diet lose this protection. An epidemiology report showed the importance of the adolescent period in humans, demonstrating a significant reduction of breast cancer risk in 13-15year old adolescent girls consuming soy.28 Genistein has been reported to be an anti-oxidant, to inhibit protein tyrosine kinases, topoisomerase II and angiogenesis, and to induce cell differentiation.29
Over the last decade, an emphasis has been placed on investigating the role of environmental chemicals on puberty and predisposition for breast cancer. Importantly, the mammary gland in humans is, to a great extent postnatally developed and subject to many endogenous and exogenous stimuli. With this comes the need to implement new technology to measure biomarkers of effect and susceptibility for cancer in order to determine if early chemical exposure can predispose to breast cancer. This report breaks from the accepted dogma of using genomic markers and moving to a more practical aspect of biomarkers that actually reflect function, proteins. Proteins, as enzymes, cofactors and regulators, actually carryout the enzymatic actions and support many metabolic processes. Although there are a plethora of papers that examine gene expression, the latter may not always translate into protein action. Working with collaborative teams of basic scientists and epidemiologists from several institutions, we have teamed up to optimized our resources (human recruitments, animal models and technology) to investigate protein biomarkers of effect from prepubertal girls from whom blood and urine were collected. From the urine of these girls, the concentration of chemical analytes was available.30 From our animal studies, we determined that two of the measured chemicals, BPA and genistein, could alter predisposition for mammary cancer in animal models.31 Two other chemicals, DEHP and BBP, were also of interest to the group because of their high metabolite concentrations in the urine of these young girls. Using recently optimized methods of rat blood protein enrichment and tagging, and mass spectrometry analysis for identification of proteins,32 we report for the first time blood protein biomarkers of effect and susceptibility from prepubertal girls with high concentrations of BPA, genistein, MEHP and MBzP (the latter two, metabolites of DEHP and BBP, respectively).
Chemicals
Rabbit polyclonal antibodies to endothelium-converting enzyme (ECE-1; cat# sc-25841) and deleted in liver cancer 1 (DLC-1; cat# sc-32931) were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX). Rabbit monoclonal antibody to eukaryotic initiation factor 3a (EIF-3a; cat# 3411) was purchased from Cell Signaling Technology (Danvers, Massachusetts). The secondary antibody (Goat anti-Rabbit IgG HRP Affinity Purified PAb) was purchased from R&D Systems, Inc. (Minneapolis, MN) and Chemiluminescent Substrate from Thermo Fisher Scientific Inc. (Rockford, IL). SepproR IgY-H7 human-specific spin columns and BPA were purchased from Sigma Chemical Co., St. Louis MO, and TMT label reagent from Thermo Scientific, Lafayette, CO. Genistein was provided by DSM Nutritional Products (Basel, Switzerland).
Human blood amples
This Genes, Environment, and Health Initiative study utilized blood collected from girls recruited via an epidemiology component of a Breast Cancer and Environment Research Center study.33 The latter was a longitudinal study of girls enrolled at 6-7years of age and followed through puberty. Enrollment occurred during 2004-2007 at Cincinnati Children’s Hospital, which recruited in the greater Cincinnati, OH metropolitan area. Informed consent from parent or guardian was obtained with child assent; the study was approved by the Institutional Review Board at Cincinnati Children’s Hospital Medical Center. Eligibility included age (six to seven years old), female, and no underlying endocrine medical conditions. BPA and genistein, and the metabolites, MEHP for DEHP, and MBzP for BBP were measured from urine samples of girls who were prepubertal at ages six and seven. Details of the procedures used for the measurement of these environmental chemical analytes were provided elsewhere.30 These measurements were used to separate the girls into quintiles of exposure for the above listed chemicals. Girls with urine creatinine-corrected concentrations in the top quintile for a single chemical, but having urine creatinine-corrected concentrations below the median for all of the other chemicals, were selected for this study. Due to the limited number of blood samples available, samples were pooled as described below. Nine blood samples from girls in the top quintile of BPA exposure were pooled to create three high BPA (H-BPA) samples. Six blood samples from girls in the top quintile of MBzP exposure were pooled to create three H-MBzP samples. The three H-MEHP and three H-genistein samples were not pooled. Six blood samples from the girls with urine concentrations below the median for all chemicals studied were pooled to create three control samples.
Serum sample preparation and at acquisition
Three pooled serum samples for each group (controls, H-BPA, H-MEHP, H-MBzP, H-genistein) were depleted of the seven most abundant proteins via SepproR IgY-H7 human specific spin columns, concentrated via molecular weight cut off centrifugal filter devices, and protein concentrations of the concentrated samples were determined with the Bradford protein assay (Bio-Rad, Hercules, CA). One hundred micrograms of protein from each group were labeled with Amine-Reactive Tandem Mass Tag Reagents (TMT 6 Label Reagents; Thermo Scientific, Lafayette, CO) according to the protocol supplied by the manufacturer as previously described.32 Briefly, protein was solubilized in 100mM TEAB plus 0.1% SDS and reduced with 9.5mM tris(2-carboxyethyl)phosphine for 1 hour at 55°C and then alkylated with 17mM iodoacetamide for 30min in the dark, and digested with Trypsin Gold overnight at 37°C (Promega, Madison WI). Each sample was incubated with a specific TMT tag reconstituted in 41μL of AcN for 1 hour at room temperature. The reaction was quenched by adding 8μL of 5% hydroxylamine. Tagged samples were combined, and the labeled peptides were purified using a SCX Macrotrap (Cat.#TR1/25109/55, Michrom Bioresources Inc. Auburn CA), and desalted using a Peptide Macrotrap (Cat.#TR1/25109/52, Michrom Bioresources Inc.). Sample volumes were reduced in a Speed Vac to near dryness, and resuspended in 95% ddH2O/ 5% ACN/ 0.1% formic acid to give a concentration of 2.5μg/μl.
Tagged samples were processed for analysis via an LTQ XL ion trap mass spectrometer equipped with a nano-electrospray source, and a Surveyor plus binary high-pressure liquid chromatography (HPLC) pump (Thermo Scientific, San Jose, CA) using a split flow configuration. Separations were carried out using a 14-fraction MudPIT approach, where the first column was a double-fritted 150micron IDx7cm SCX (Poly SULFOETHYL A 300 A, 5 micron, PolyLC), connected to a 150micronx13cm pulled tip C-18 column (Jupiter C-18 300 A, 5 micron, Phenomenex, Torrance, CA). The HPLC was set up with two mobile phases that included solvent A (0.1% formic acid in ddH2O), and solvent B (0.1% formic acid in 85% ddH2O/15% ACN), and was programmed as follows; 15min @ 0%B (2µL/min, load), 65min @ 0%-50%B (~0.5nL/min, analyze) and 20min @ 0%B (2µL/min, equilibrate). This gradient was used for each step of the MudPIT analysis, in which the flow-through was first analyzed, followed by 13 additional fractions obtained by 35μL injections of the following concentrations of ammonium acetate dissolved in ddH2O: 25mM, 32.5mM, 40mM, 50mM, 75mM, 100mM,150mM, 200mM, 250mM, 300mM, 350mM, 400mM, and 1M. The LTQ XL was operated in data dependent triple play mode, with a survey scan range of 350-2000m/z, followed by a zoom scan for charge state determination, and pulsed Q dissociation (PQD) scan for MS2, which were carried out with 2.0 Da isolation widths on the 3 top most intense ions. MS data were collected in profile mode for all scan types. Charge state screening and data dependent dynamic exclusion were enabled, with exclusion of non-assigned peptides, aminimum signal intensity of 2000, a repeat count of 2, and exclusion duration of 90s for ions +/- 1.5m/z of the parent ion. The automatic gain control (AGC) settings were 3X104, 5x103, and 5x104 ions for survey, zoom, and PQD modes respectively. Scan times were set at 25, 50, and 250ms for survey, zoom, and PQD modes respectively. For PQD, the activation time, activation Q, and normalized collision energy were set at 0.1ms, 0.7, and 35% respectively. The spray voltage was set at 1.9kV, with a capillary temperature of 170°C. All Mud PIT runs were carried out in duplicate.
Data analysis
The XCalibur RAW files were centroided and converted to MzXML and themgf files were then created using both ReAdW and MzXML2Search respectively (http://sourceforge.net/projects/sashimi/). The data was searched using SEQUEST (v.27 rev12, .dta files), set for two missed cleavages, a precursor mass window of 0.45 Da, tryptic enzyme, variable modification M @ 15.9949, and static modifications C @ 57.0293, K and N-term@ 229.1629. Searches were performed with a rat subset of the UniRef100 database, which included common contaminants such as digestion enzymes and human keratins. Identified peptides were filtered, grouped, and quantified using ProteoIQ (Premierbiosoft, Palo Alto, CA). Only peptides with charge state of ≥2+, aminimum peptide length of 6 amino acids, and non-zero quantities for all six mass tags were accepted for analysis. ProteoIQ incorporates the two most common methods for statistical validation of large proteome datasets, false discovery rate (FDR), and protein probability.34,35 The FDR was set at <1% cut-off, with a total group probability of ≥0.7, with at least 2 peptides assigned per protein. Relative quantification was performed via spectral counting, and spectral count abundances were normalized between samples.
Animals
Cross–species and tissue validation of selected proteins were carried out from mammary tissue extracts of rats exposed prepubertally to BPA and genistein. Animal studies were conducted in accordance with the University of Alabama at Birmingham Guidelines for Animal Use and Care. Seven week old female Sprague-Dawley rats were purchased from Charles River Laboratories (Wilmington, MA). Animals were housed in a temperature controlled facility with a 12-h light: dark cycle. These animals were bred with proven Sprague-Dawley studs in our facilities, provided phytoestrogen-free AIN-76A diet (Harlan Teklad, Madison, WS) and water via glass bottles, and housed in polypropylene cages (all polycarbonate/BPA free). On theday of birth, offspring were sexed, and litters were culled to 10 offspring per lactating dam. For BPA treatment, lactating dams were gavaged intragastrically with 250μg BPA/kg/day while controls received an equivalent volume of the vehicle sesame oil beginning on postnatal day 2 (PND2) and continuing through PND20. For genistein treatment, the lactating dams received 250mg genistein/kg AIN-76A diet or AIN-76A diet only as controls from PND2 through PND20. In this manner, the offspring are exposed to genistein or BPA via the mother’s milk. Offspring were weaned on PND21 and continued on AIN-76A diet only. At PND50, female offspring were killed in the estrous phase. The fourth abdominal mammary glands were rapidly dissected from live ketamine/xylazine anesthetized animals (tominimize proteolysis), snap-frozen in liquid nitrogen, and stored at -80ºC for later analysis.
Western blot validation
Western blot analyses were performed to validate changes in protein expressions detected by TMT-MS. Since human sera were limited, we carried out “cross-species” validation in mammary glands from 50day old rats exposed prepubertally to BPA and genistein (and controls). To determine the changes in protein expression, six mammary gland samples per treatment group were analyzed by Western blots. Each sample was derived from only one rat randomly selected from separate litters per treatment group. The same quantity of protein from each sample was separated by SDS-PAGE and transferred to a nitrocellulose membrane (Bio-Rad, Hercules, CA). The membranes were blocked and immuno blotted with appropriate antibodies including deleted in liver cancer 1 (DLC-1), endothelium-converting enzyme (ECE-1) and eukaryotic initiation factor 3 (EIF3). Molecular weight ladders (Bio-Rad) were used to validate the proteins of interest. Nitrocellulose membranes were incubated with an appropriate secondary antibody conjugated to HRP (R&D Systems, Inc) followed with a chemiluminescent substrate and exposed to X-ray radiography film. Quantitative analysis of protein expression was accomplished by scanning autoradiogram and densitometry (Image J, NIH).Western blot analyses were performed to validate changes in protein expressions detected by TMT-MS. Since human sera were limited, we carried out “cross-species” validation in mammary glands from 50day old rats exposed prepubertally to BPA and genistein (and controls). To determine the changes in protein expression, six mammary gland samples per treatment group were analyzed by Western blots. Each sample was derived from only one rat randomly selected from separate litters per treatment group. The same quantity of protein from each sample was separated by SDS-PAGE and transferred to a nitrocellulose membrane (Bio-Rad, Hercules, CA). The membranes were blocked and immunoblotted with appropriate antibodies including deleted in liver cancer 1 (DLC-1), endothelium-converting enzyme (ECE-1) and eukaryotic initiation factor 3 (EIF3). Molecular weight ladders (Bio-Rad) were used to validate the proteins of interest. Nitrocellulose membranes were incubated with an appropriate secondary antibody conjugated to HRP (R&D Systems, Inc) followed with a chemiluminescent substrate and exposed to X-ray radiography film. Quantitative analysis of protein expression was accomplished by scanning autoradiogram and densitometry (Image J, NIH).
Statistical analysis
For urinary metabolite concentrations, each pooled sample group was compared against prepubertal control using a T-test for unequal variance to determine the statistical significance of the difference in means of the top quintile and control groups. P-values of <0.05 were considered statistically significant. For the proteomic data generated by TMT-MS, separate non-parametric statistical analyses were performed between each set of comparison groups. These non-parametric analyses include 1) the calculation of weight values by significance analysis of microarray34,35 (SAM; cut off |}0.8|combined with 2) Wilcoxon (cut off of p<0.05) which then were sorted according to the highest statistical relevance in each comparison. For SAM, whereby the weight value (W) is a statistically derived function that approaches significance as the distance between the means (μ1-μ2) for each group increases, and the SD (δ1-δ2) decreases using the formula, W=(μ1-μ2)/( δ1+δ2). For protein abundance ratios determined with TMT-MS, we set a 2.0 fold change as the threshold for significance, determined empirically by analyzing the inner-quartile data from the control experiment indicated above using ln-ln plots, where Pierson’s correlation coefficient (R) was 0.98, and >99% of the normalized intensities fell between +/-1.5 fold. In each case, any two of the three tests (SAM, Wilcoxon, or fold change) had to pass. Statistical analysis of Western blot analysis was performed by using one way analysis of variance (ANOVA) to determine significance (P<0.05).
Bioinformatics and systems biology analysis
Those proteins found to have significantly changed in the high chemical exposed groups versus the control group were evaluated for biological significance, and for key biological functions to cancer related pathways. PANTHER (Protein Analysis through Evolutionary Relationships System) (http://www.pantherdb.org) was used for protein classification. Ingenuity Pathways Analysis (Ingenuity Systems, www.ingenuity.com) (IPA, Redwood City, CA) was used to identified the pathways from the IPA library of canonical pathways that were most significant in our dataset. Proteins from the dataset that met the criteria for differential expression and were associated with a canonical pathway in the Ingenuity Knowledge Base were considered for the analysis. Finally, proteins of interest were subjected to literature searches as to biological function and appropriateness as biomarkers of cancer susceptibility.
The group of girls with high urinary concentrations of BPA, MEHP, and MBzP were significantly increased compared against respective controls (defined as urine samples below the median of the chemicals analyzed) (Table 1). While girls with high urinary concentrations of genistein were increased 38 fold as compared to control urine samples, these did not reach statistical significance.
Biomarker |
Group |
N |
Mean ± SD |
P-Value |
BPA |
Control |
6 |
1.1 ± 0.4 |
|
|
H-BPA |
9 |
17.5 ± 11.2 |
p<0.01 |
Genistein |
Control |
6 |
33.9± 20.9 |
|
|
H-Genistein |
3 |
1286.4 ± 860.3 |
p=0.13 |
MEPH |
Control |
6 |
1.6± 0.9 |
|
|
MEHP |
3 |
17.2 ± 0.7 |
p<0.0001 |
MBzP |
Control |
6 |
14.4 ± 8.2 |
|
|
MBzP |
6 |
148.5 ± 119.4 |
p<0.05 |
Table 1 Urinary Creatinine (Cr) Corrected Biomarker Concentrations (ng/g Cr)*
*Creatinine corrections were determined using creatinine measurements from the same urine specimens as for the blood biomarker measurements. A T-test for unequal variance was used to determine the statistical significance of the difference in means of the top quintile and control groups. BPA: Bisphenol A; MEHP: mono-ethyl hexyl-phthalate; MBzP: mono-benzyl phthalate.
Systems biology analysis
Using TMT-MS technology, we identified 51, 34, 57 and 47 proteins to be differentially expressed from the blood of girls with H-BPA, H-genistein, H-MEHP and H-MBzP urine concentrations, respectively (Tables 2-5). Analysis of biological function via PANTHER demonstrates that all four exposure groups had similar responses for metabolic process (26-28%), cellular process (21-23%), biological regulation (10-12%) and developmental process (7-9%) (Figure 1). However, H-BPA and H-genistein exposure groups had higher responses on localization (13% and 14% responses, respectively) than the H-MEHP and H-MBzP groups (4% and 8%, respectively). In regard to cellular component organization or biogenesis, the H-genistein group had only 5% response while H-BPA, H-MEHP and H-MBzP groups had approximately 10% responses. Multicellular organismal process accounted for 7% of biological function in H-BPA and H-genistein girls, and 4% and 1% in H-MEHP and H-MBzP girls, respectively. Proteins associated with reproduction were highest in MEHP girls (7%), and apoptotic process and biologic adhesion constituted 3% of biological functions in H-genistein and H-MBzP girls.

Of the 189 total proteins identified in the four groups of girls, only one (E3 ubiquitin-ligase) was up regulated in the blood of both H-BPA and H-MEHP girls. E3 ubiquitin-ligases have been reported to promote cancer cell proliferation and are frequently over expressed in human cancer development.36 They are also associated with chemo-resistance, poor clinical prognosis, and have been suggested as potential cancer biomarkers. On the other hand, cycilin-1, a protein reported to play a role in spermatid differentiation,37 was down regulated in H-MEHP girls and up regulated in H-MBzP girls. The significance of cycilin-1 in girls has not yet been identified. Beyond these two proteins, we noted a large number of unique protein signatures in the blood of each group. We expected, and found, that these exposure groups yielded different biomarkers of effect. Therefore, we assessed individual protein function and canonical pathway analysis towards cancer for all four chemical exposure groups, followed this up with literature searches, and considered directional change in protein expressions.
Protein IDa |
Protein Nameb |
Group Prob-ability |
No. Unique Peptides |
Fold Change (Rx/C)c |
SAMd |
P10323 |
acrosin |
0.99 |
4 |
+2.80 |
1.48 |
O75969 |
A-kinase anchor |
0.94 |
2 |
+2.52 |
2.38 |
Q01484 |
ankyrin-2 |
0.99 |
4 |
+2.21 |
1.69 |
P46013 |
antigen Ki-67 |
0.95 |
4 |
+3.65 |
1.41 |
Q9P203 |
BTB/POZ domain-containing 7 |
0.94 |
3 |
+2.52 |
2.09 |
Q9UPV0 |
centrosomal protein of 164 |
0.98 |
2 |
+3.04 |
1.84 |
B7ZML1 |
chromodomain helicase DNA binding protein |
0.82 |
4 |
+3.06 |
1.68 |
Q5I2W7 |
cytochrome P450, family 24, A1 |
0.72 |
2 |
-2.57 |
1.16 |
Q8TDD1 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 |
0.87 |
4 |
-2.17 |
1.09 |
Q5T1A1 |
DC-stamp domain-containing 2 |
0.93 |
3 |
+2.33 |
1.07 |
Q96QB1 |
deleted in liver cancer 1 (DLC1) |
0.83 |
3 |
-2.95 |
1.24 |
Q9NZW4 |
dentin sialophosphoprotein |
0.99 |
4 |
-2.20 |
1.00 |
Q9UBC3 |
DNA (cytosine-5)-methyltransferase 3B (DNMT3B) |
0.93 |
2 |
-2.51 |
1.77 |
P11532 |
dystrophin |
0.92 |
4 |
+3.18 |
0.99 |
B4E391 |
elongation factor Ts |
0.98 |
2 |
-4.37 |
1.37 |
Q9UPY3 |
endoribonuclease dicer |
0.80 |
2 |
+2.00 |
1.56 |
O60841 |
eukaryotic translation initiation factor 5B |
0.99 |
4 |
+2.19 |
1.00 |
Q9NV70 |
exocyst complex component |
0.95 |
3 |
+2.02 |
2.32 |
Q149M6 |
exophilin 5 |
1.00 |
4 |
+2.79 |
1.72 |
Q8IYW5 |
E3 ubiquitin- ligase |
0.96 |
2 |
+2.45 |
1.00 |
O14964 |
hepatocyte growth factor-regulated tyrosine kinase substrate |
0.90 |
2 |
+2.48 |
0.87 |
Q7Z2Y8 |
interferon-induced very large GTPase 1 |
0.83 |
2 |
+4.50 |
0.89 |
A6PVS8 |
leucine-rich repeat and IQ domain-containing 3 |
0.89 |
2 |
+2.33 |
0.84 |
Q8NEZ4 |
lysine-specific methyltransferase 2C |
0.99 |
6 |
+2.44 |
0.81 |
O14686 |
lysine-specific methyltransferase 2D |
0.94 |
3 |
+2.02 |
1.62 |
Q8N6Q8 |
methyltransferase-like protein 25 |
0.91 |
4 |
-2.52 |
1.48 |
O60307 |
microtubule-associated serine/thr- kinase 3 |
0.84 |
3 |
+3.48 |
1.24 |
P45985 |
mitogen-activated kinase kinase 4 (MKK4) |
0.95 |
2 |
+3.08 |
1.12 |
Q7Z4B4 |
MSTP159 |
0.97 |
2 |
+6.14 |
0.97 |
P12882 |
myosin-1 a, b |
0.99 |
5 |
-2.13 |
0.80 |
Q9UKX2 |
myosin-2 a,,b |
0.91 |
2 |
-2.13 |
0.80 |
Q9Y4I1-3 |
myosin-va |
0.98 |
5 |
+3.13 |
1.11 |
P14543 |
nidogen _1 |
0.98 |
2 |
+2.31 |
0.81 |
P49790 |
nucleoporin 153 |
0.74 |
3 |
-2.08 |
1.66 |
Q8IVF1 |
NUT family member 2A |
0.97 |
3 |
-3.44 |
1.62 |
A6NNL0 |
NUT family member 2B |
0.98 |
3 |
+3.29 |
0.91 |
Q9HC10 |
otoferlin |
0.84 |
2 |
+2.30 |
2.79 |
Q92576 |
PHD finger 3 |
0.79 |
2 |
-4.13 |
1.38 |
B7Z9C9 |
potassium large conductance ca-activated channel, M beta 3 |
0.96 |
2 |
+2.13 |
0.97 |
Q6NUQ1 |
RAD50-interacting 1 |
0.92 |
2 |
-2.68 |
1.05 |
P49247 |
ribose-5-phosphate isomerase |
0.94 |
2 |
-3.61 |
0.88 |
Q92730 |
rho family GTPase 1 |
0.94 |
2 |
-3.61 |
0.88 |
Q9Y4G6 |
talin-2 |
0.90 |
4 |
+2.54 |
1.43 |
Q16881 |
thioredoxin reductase 1 |
0.92 |
3 |
+2.19 |
1.21 |
Q00WD0 |
THO complex subunit 4 |
0.80 |
3 |
-2.06 |
2.15 |
Q12931 |
TNF receptor-associated protein 1 |
0.81 |
2 |
+2.51 |
1.01 |
Q9Y2W1 |
thyroid hormone receptor-associated protein 3 |
0.90 |
4 |
+4.30 |
4.04 |
Q9UL62 |
transient receptor potential channel 5 (TRPC5) |
0.93 |
3 |
+2.40 |
1.23 |
P17948 |
vascular endothelial growth factor receptor 1 |
1.00 |
4 |
-2.21 |
0.87 |
Q64LD2 |
WD repeat-containing 25 |
0.99 |
2 |
+2.25 |
0.86 |
O15231 |
zinc finger 185 |
0.92 |
2 |
+2.72 |
0.98 |
Table 2 Differentially Regulated Proteins Identified by TMT-MS in Sera of Girls with High Urine Concentrations of BPA
aProteins identified using the Human Genome DB ID.
bProteins in bold are carcinogenesis-related proteins.
cPositive and negative fold change in protein expression indicate up- and down-regulation of protein expression relative to control, respectively.
dSignificance analysis of microarray (SAM); cut off |±0.8| calculated as described in Materials and Methods.
High-BPA group
From the H-BPA group of biomarkers of effect, IPA canonical pathways of function identified 10 proteins to be associated with cancer, seven of these being up regulated: ankyrin 2, antigen Ki-67, E3 ubiquitin-ligase, talin 2, transient receptor potential channel 5 (TRPC5), mitogen-activated kinase kinase 4 (MKK4) and zinc finger 185 (Table 2). Ankyrin 2 is a cytoskeletal protein that plays a role in metastatic breast tumor cell invasion and migration.38 Antigen Ki-67 is a cellular marker for proliferation and used as a biomarker for cancer.39 Furthermore, we have previously reported that exposure of rats to BPA resulted in increased Ki67 protein expression in mammary glands40 and increased chemically-induced mammary cancer in rats.13 As noted in the previous paragraph, up regulated E3 ubiquitin-ligase has been reported to play a role in carcinogenesis and proposed as a biomarker of cancer.36 Talin2 is up regulated in invasive breast carcinomas.41 The expression of TRPC5 protein is high in human breast tumors and in the circulation.42 MKK4 is an important mediator of cellular responses to extracellular signals that include growth factors, hormones, cytokines and environmental stresses.43 The role of MKK4 in cancer development appears complex, as some studies support pro-oncogenic mechanisms, while others suggest suppressor protein action.44 The JNK and p38 pathways are implicated in tumor suppression in the presence of loss of function mutations in the MKK4 gene,45,46 while MKK4 and JNK can participate in tumor formation, suggesting a more complex role for this pathway in tumor development.45,47 While it is plausible that up regulated MKKA could be a biomarker of BPA chemical exposure, the partnering of MKK4 with other signaling proteins appears to determine the functional outcome. On the other hand, up regulation of Zinc finger 185 in H-BPA girls is not consistent with carcinogenesis. Zinc finger 185 has been reported to function as a growth inhibitory protein by associating with the actin–cytoskeleton.48 But, up regulated zinc finger 185 in girls with H-BPA does not support the concept of this protein contributing to cancer causation.
Protein IDa |
Protein Nameb |
Group Prob-ability |
No. Unique Peptides |
Fold Change (Rx/C)c |
SAMd |
Q9H0C2 |
ADP/ATP translocase 4 |
0.99 |
3 |
+2.02 |
1.15 |
A4D0V7 |
cadherin-like and PC-esterase domain containing 1 |
0.95 |
2 |
-2.21 |
1.22 |
Q8WW4 |
chromosome 2 Open Reading Frame 47 |
0.93 |
2 |
-4.56 |
0.91 |
A6NNT2 |
chromosome 16 open reading frame 96 |
0.95 |
3 |
-2.07 |
5.27 |
P53621 |
coatomer alpha subunit |
0.99 |
5 |
-2.18 |
1.10 |
O43293 |
death associated kinase |
0.86 |
3 |
-2.77 |
1.38 |
P52429 |
diacylglycerol kinase epsilon |
0.90 |
2 |
-3.25 |
1.29 |
Q9C098 |
doublecortin-like kinase 3 |
0.98 |
4 |
-2.66 |
1.40 |
P42892 |
endothelin-converting enzyme (ECE-1) |
1.00 |
4 |
-2.12 |
1.72 |
B4DUI3 |
eukaryotic translation initiation factor 3 subunit J (EIF3) |
0.82 |
2 |
-2.30 |
1.63 |
Q96JP0 |
fem-1 homolog C |
0.92 |
2 |
+2.02 |
1.45 |
Q5CZC0 |
fibrous sheath-interacting 2 |
0.93 |
5 |
-2.17 |
0.97 |
O60318 |
germinal-center associated nuclear protein |
0.88 |
2 |
-2.26 |
1.09 |
B9A064 |
immunoglobulin lambda-like polypeptide 5 |
0.92 |
3 |
-3.48 |
1.11 |
Q6NSI8 |
KIAA1841 |
0.98 |
3 |
-2.70 |
1.47 |
Q5QGS0 |
KIAA2022 |
0.91 |
2 |
+2.56 |
1.13 |
Q9Y496 |
kinesin-like protein |
0.82 |
2 |
+2.47 |
1.20 |
Q9UMY1 |
nucleolar 7 |
0.82 |
2 |
+2.10 |
1.02 |
Q9NQX1 |
PR domain zinc finger 5 (PRDM5) |
0.97 |
5 |
+2.30 |
0.95 |
Q9UQ6 |
regulating synaptic membrane exocytosis 2 |
0.94 |
3 |
-2.82 |
0.93 |
A8K0H3 |
ribosomal L29 |
0.99 |
3 |
-2.73 |
1.81 |
Q8IYF1 |
RNA polymerase II transcription factor SIII sub A2 |
0.89 |
4 |
+2.28 |
1.22 |
P10523 |
s-arrestin |
0.96 |
2 |
-2.38 |
3.26 |
Q6R2W3 |
SCAN domain-containing 3 |
0.93 |
2 |
-2.27 |
0.84 |
A1X283 |
SH3 and PX domain-containing 2B |
0.78 |
2 |
-2.79 |
1.46 |
Q7Z5N4 |
sidekick |
0.83 |
3 |
-2.31 |
1.68 |
Q9P2W9 |
syntaxin-18 |
0.94 |
2 |
+2.31 |
1.32 |
Q8N3R3 |
T-Cell activation inhibitor |
0.77 |
2 |
+4.56 |
1.51 |
O75643 |
U5 small nuclear ribonucleo 200 kDa helicase |
0.97 |
4 |
-2.11 |
1.19 |
Q6IE27 |
vitellogenin-like 1 |
0.97 |
4 |
+2.48 |
1.73 |
Q8IWG1 |
WD repeat-containing protein 63 |
0.96 |
3 |
-6.05 |
1.02 |
Q96DT7 |
zinc finger and BTB domain-containing 10 |
0.89 |
3 |
-4.31 |
0.85 |
B4DSE6 |
zinc finger 485 |
0.92 |
2 |
-2.76 |
0.89 |
Q8N49 |
zinc finger 808 |
0.85 |
3 |
-3.05 |
2.62 |
Table 3 Differentially Regulated Proteins Identified by TM-MS in Sera of Girls with High Urine Concentrations of Genistein
aProteins identified using the Human Genome DB ID.
bProteins in bold are consistent with cancer prevention.
cPositive and negative fold change in protein expression indicate up- and down-regulation of protein expression relative to control, respectively.
dSignificance analysis of microarray (SAM); cut off |±0.8| calculated as described in Materials and Methods.
Protein IDa |
Protein Nameb |
Group Prob-ability |
No. Unique Peptides |
Fold Change (Rx/C)c |
SAMd |
Q9H6R3 |
acyl-CoA synthetase short-chain family member 3 |
0.91 |
2 |
+2.57 |
1.65 |
Q9BXX2 |
ankyrin repeat domain-containing 30B |
0.97 |
4 |
+2.16 |
1.44 |
Q8N7J2 |
APC membrane recruitment protein 2 |
0.98 |
3 |
-2.80 |
0.84 |
A8Y988 |
ATP-binding cassette (Sub-family C, member 6) |
1.00 |
3 |
-2.21 |
1.40 |
Q8TD16 |
bicaudal D homolog 2 |
0.93 |
2 |
-2.70 |
1.57 |
P27815 |
cAMP-specific 3',5'-cyclic phosphodiesterase 4A |
0.95 |
5 |
-3.11 |
1.28 |
P14384 |
carboxypeptidase M |
0.95 |
2 |
-2.09 |
1.01 |
Q86X52 |
chondroitin sulfate synthase 1 |
0.98 |
3 |
-2.17 |
1.54 |
Q9P2D1 |
chromodomain-helicase-DNA-binding 7 |
1.00 |
5 |
-2.36 |
1.69 |
Q9HCK8 |
chromodomain-helicase-DNA-binding 8 |
0.95 |
5 |
-2.70 |
1.81 |
Q96N11 |
chromosome 7 open reading frame 26 |
0.83 |
3 |
-2.41 |
1.94 |
A6PVK7 |
chromosome 9 open reading frame 84 |
1.00 |
3 |
-2.26 |
0.90 |
Q96PX6 |
coiled-coil domain-containing 85A |
0.92 |
4 |
+2.15 |
1.46 |
Q8IYT3 |
coiled-coil domain containing 170 |
0.98 |
2 |
+2.39 |
0.87 |
P0C0L5 |
complement C4B |
1.00 |
3 |
+2.58 |
0.80 |
D3DUT5 |
complement component 1 |
1.00 |
3 |
-2.02 |
1.51 |
Q9NX05 |
constitutive coactivator of PPAR-gamma-like 2 |
0.81 |
2 |
+3.51 |
1.27 |
Q9NXE8 |
CWC25 spliceosome-associated protein homolog |
0.86 |
2 |
-3.38 |
1.83 |
P35663 |
cylicin-1 |
0.95 |
3 |
-3.44 |
1.14 |
Q14008 |
cytoskeleton-associated 5 |
0.97 |
4 |
-2.73 |
4.09 |
Q8N254 |
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 1 |
0.85 |
3 |
-2.47 |
0.85 |
Q8N9I9-2 |
deltex 3, E3 ubiquitin ligase |
0.97 |
3 |
-2.43 |
0.97 |
Q9UN19 |
dual adapter for phosphotyrosine and 3-phosphoinositides |
0.89 |
2 |
-2.16 |
0.89 |
Q8WV44 |
E3 ubiquitin- ligase |
0.81 |
2 |
+3.06 |
0.81 |
Q96RP9-2 |
elongation factor G |
0.89 |
3 |
-7.97 |
0.89 |
Q13003 |
glutamate receptor, ionotropic kainate 3 |
1.00 |
3 |
-3.36 |
1.00 |
P14866 |
heterogeneous nuclear ribonucleo L |
0.78 |
2 |
-2.45 |
0.78 |
P42858 |
huntingtin |
0.89 |
2 |
+2.43 |
0.89 |
P17936 |
insulin-like growth factor-binding protein 3 (IGFBP-3) |
1.00 |
3 |
-3.44 |
1.00 |
Q14624 |
inter-alpha-trypsin inhibitor heavy chain H4 |
0.99 |
4 |
-2.17 |
0.99 |
Q9HCM1 |
KIAA1551 |
0.99 |
6 |
-2.79 |
0.99 |
Q9C099 |
leucine-rich repeat and coiled-coil domain-containing 1 |
0.98 |
5 |
-2.19 |
0.98 |
Q9NZR2 |
low-density lipo receptor-related 1B |
0.96 |
3 |
-2.20 |
0.96 |
P26927 |
macrophage stimulating protein 1 |
1.00 |
3 |
-2.05 |
1.00 |
Q8NB16 |
mixed lineage kinase domain-like |
0.95 |
2 |
-3.58 |
0.95 |
Q96T76-7 |
MMS19 nucleotide excision repair homolog |
0.85 |
2 |
-2.51 |
0.85 |
P11055 |
myosin-3 |
0.83 |
4 |
-2.37 |
0.83 |
Q3T906 |
N-acetylglucosamine-1-phosphotransferase subunits alpha/beta |
0.98 |
3 |
-2.41 |
0.98 |
P37198 |
nuclear pore glyco p62 |
0.90 |
2 |
-3.00 |
0.90 |
Q17RR3 |
pancreatic lipase-related 3 |
0.98 |
2 |
-3.20 |
0.98 |
O00329 |
phosphatidylinositol 3-kinase delta catalytic subunit |
0.77 |
2 |
-2.07 |
0.77 |
Q86UU1 |
pleckstrin homology-like domain family B member 1 |
1.00 |
4 |
-2.24 |
1.31 |
Q4G0U5 |
primary ciliary dyskinesia 1 |
0.96 |
2 |
+3.59 |
2.96 |
O95206 |
protocadherin-8 |
1.00 |
3 |
-5.00 |
1.25 |
B7Z4Q3 |
RNA-binding Luc7-like |
0.82 |
3 |
-2.03 |
0.84 |
Q6PCB5 |
round spermatid basic 1-like |
0.98 |
3 |
-2.19 |
1.24 |
B4DZM2 |
SFRS2-interactin |
1.00 |
6 |
-2.02 |
2.18 |
Q59HF0 |
solute carrier family 13 (sodium/sulfate symporters), member 4 |
0.93 |
2 |
-3.01 |
0.81 |
B7ZLS9 |
synaptonemal complex 1 |
1.00 |
5 |
+2.26 |
1.03 |
O15061 |
synemin |
1.00 |
5 |
-2.32 |
2.79 |
A0AVF1 |
tetratricopeptide repeat 26 |
0.88 |
2 |
-2.88 |
0.92 |
Q9BUB4 |
tRNA-specific adenosine deaminase 1 |
0.97 |
3 |
-3.73 |
1.04 |
Q2NL82 |
TSR1, 20S rRNA accumulation, homolog |
0.92 |
2 |
-2.98 |
1.60 |
Q9C0G0 |
zinc finger 407 |
0.98 |
5 |
-2.30 |
1.10 |
A7MD06 |
zinc finger 483 |
0.91 |
2 |
-2.02 |
1.86 |
Q9NUD5 |
zinc finger CCHC domain-containing 3 |
0.97 |
2 |
-2.17 |
1.06 |
Q9CX48 |
zinc finger CCHC domain-containing 10 |
0.73 |
3 |
-2.98 |
1.03 |
Table 4 Differentially Regulated Proteins Identified by TMT-MS in Sera of Girls with High Urine Concentrations of MEHP
aProteins identified using the Human Genome DB ID.
bProteins in bold are carcinogenesis-related proteins.
cPositive and negative fold change in protein expression indicate up- and down-regulation of protein expression relative to control, respectively.
dSignificance analysis of microarray (SAM); cut off |±0.8| calculated as described in Materials and Methods.
Protein IDa |
Protein Nameb |
Group Prob-ability |
No. Unique Peptides |
Fold Change (Rx/C)c |
SAMd |
Q9UJX4 |
anaphase-promoting complex subunit 5 |
0.87 |
2 |
-2.26 |
1.26 |
P02649 |
apolipo E |
0.99 |
2 |
+2.16 |
0.82 |
Q9UBJ2 |
ATP-binding cassette sub-family D member 2 |
0.86 |
2 |
-3.45 |
3.18 |
Q5XSY0 |
ATP synthase subunit a |
0.96 |
2 |
+2.25 |
1.09 |
Q9NPI1 |
bromodomain-containing 7 |
1.00 |
3 |
+2.47 |
0.90 |
O14578 |
citron Rho-interacting kinase |
0.99 |
5 |
+2.95 |
1.41 |
P35663 |
cylicin-1 |
0.99 |
4 |
+2.16 |
1.02 |
Q9HCK1 |
DBF4-type zinc finger-containing 2 |
1.00 |
5 |
-2.34 |
1.10 |
O60231 |
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
0.88 |
3 |
+4.94 |
1.53 |
Q99543 |
DnaJ homolog subfamily C member 2 |
0.99 |
2 |
+2.57 |
0.92 |
O60673 |
DNA polymerase zeta catalytic subunit |
0.92 |
2 |
-2.66 |
0.95 |
Q8WXX0 |
dynein heavy chain 7 |
0.80 |
2 |
+2.11 |
2.33 |
O75411 |
Krueppel-like factor 10 (KLF10) |
0.95 |
2 |
+2.61 |
1.47 |
P08F94 |
fibrocystin |
0.98 |
2 |
-2.16 |
1.07 |
Q5TBA9 |
furry homolog |
0.96 |
5 |
+2.51 |
1.02 |
A6NFK2 |
glutaredoxin domain-containing cysteine-rich 2 |
0.99 |
2 |
-2.65 |
0.87 |
P10144 |
granzyme B |
0.81 |
3 |
+3.41 |
2.18 |
B4DRD6 |
histone H1 |
1.00 |
4 |
+2.28 |
1.22 |
Q9BYW2 |
histone-lysine N-methyltransferase |
0.99 |
3 |
-3.36 |
1.53 |
D3DW85 |
iron-responsive element binding 2 |
0.94 |
2 |
+2.30 |
1.47 |
O60738 |
KB07 |
0.97 |
2 |
+2.58 |
1.27 |
B0YJ32 |
laminin alpha-3 chain 1 |
1.00 |
7 |
+2.73 |
0.85 |
O95447 |
leber congenital amaurosis 5-like |
0.88 |
2 |
-3.05 |
1.84 |
B3KNZ9 |
megakaryocyte-associated tyrosine- kinase |
0.98 |
2 |
+2.37 |
1.33 |
Q6P444 |
mitochondrial fission regulator 2 |
0.98 |
2 |
+3.02 |
1.08 |
Q9Y3A3 |
MOB family member 4, phocein |
0.99 |
3 |
-2.15 |
2.70 |
Q9Y5Q3 |
musculoaponeuroticfibrosarcoma oncogene homolog B |
0.90 |
2 |
+2.17 |
1.04 |
Q9UKN7 |
myosin-XV |
0.88 |
3 |
-2.81 |
1.07 |
O60313 |
optic atrophy 1 |
0.75 |
2 |
+2.26 |
1.41 |
Q9BRX2 |
pelota homolog |
0.75 |
2 |
+2.73 |
2.16 |
Q9BVI0-2 |
PHD finger protein 20 |
0.71 |
2 |
+2.21 |
0.95 |
P78356 |
phosphatidylinositol-5-phosphate 4-kinase type-2 beta |
0.87 |
2 |
-2.80 |
1.34 |
Q9ULM0 |
pleckstrin homology domain-containing family H member 1 |
0.99 |
2 |
+2.16 |
1.02 |
Q13563 |
polycystin-2 |
0.85 |
2 |
+3.96 |
3.05 |
Q96P66 |
probable G- coupled receptor 101 |
0.84 |
2 |
+2.14 |
1.30 |
Q05BX6 |
RABEP1 |
0.91 |
3 |
+2.22 |
1.81 |
A8K322 |
RAD54 homolog B (RAD54B) |
0.85 |
3 |
+2.01 |
2.72 |
Q9NS23 |
ras association domain-containing 1 |
0.72 |
2 |
-3.29 |
1.11 |
P06400 |
retinoblastoma-associated protein (RB1) |
0.95 |
2 |
+2.30 |
0.90 |
P27169 |
serum paraoxonase/arylesterase 1 |
1.00 |
2 |
+2.61 |
2.33 |
B3KWZ6 |
sperm associated antigen 17 |
0.96 |
4 |
+2.28 |
1.01 |
Q5VWG9 |
transcription initiation factor TFIID subunit 3 |
0.80 |
2 |
+2.24 |
1.29 |
A0PJ48 |
TAOK2 |
0.97 |
4 |
+3.05 |
0.86 |
P28289 |
tropomodulin-1 |
0.93 |
2 |
+2.57 |
0.93 |
P04275 |
von Willebrand factor |
0.97 |
5 |
+2.31 |
0.97 |
O43345 |
zinc finger 208 |
0.88 |
3 |
+2.29 |
1.92 |
P49207 |
60S ribosomal L34 |
0.95 |
2 |
+2.24 |
1.31 |
Table 5 Differentially Regulated Proteins Identified by TMT-MS in Sera of Girls with High Urine Concentrations of MBzP
aProteins identified using the Human Genome DB ID.
bProteins in bold are consistent with cancer prevention.
cPositive and negative fold change in protein expression indicate up- and down-regulation of protein expression relative to control, respectively.
dSignificance analysis of microarray (SAM); cut off |±0.8| calculated as described in Materials and Methods.
Three blood proteins found by TMT-MS to be down regulated in blood of girls with high urine concentrations of BPA are deleted in liver cancer 1 (DLC1), DNA (cytosine-5)-methyltransferase 3B (DNMT3B) and RAD50-interacting 1. DLC1 functions as a tumor suppressor in a number of common cancers, including breast, liver, prostate, lung and colorectal cancers.49 Thus, a BPA mediated decrease in the tumor suppressor DLC1 may facilitate tumor development. On the other hand, DNMT3B expression is essential for mammalian development and is required for genome wide de novo methylation.50 RAD50-interacting 1 is reported to play a role in cell cycle checkpoint control after DNA damage.51 Down regulation of RAD50-interacting 1 would potentially allow cell proliferation of DNA damage to continue unchecked. In comparing the functions of the differentially expressed proteins with the directional change in protein expressions, we suggest that nine of the 10 cancer-related proteins in the blood of girls with H-BPA could actually play a role in predisposing for cancer, ankyrin-2, antigen Ki-67, E3 ubiquitin-ligase, talin-2, MKK4, TRPC5, DLC1, DNMT3B and RAD50-interacting 1.
High genistein group
In blood of girls with high genistein concentrations in their urine, two proteins with cancer associations were down regulated: endothelin-converting enzyme (ECE-1) and eukaryotic translation initiation factor 3 subunit J (EIF-3) (Table 3). ECE-1 has been implicated in the pathogenesis of a range of disease states including breast, gynecological and urological cancers, cardiovascular disease and Alzheimer’s disease.52 EIF-3 has been found elevated in human breast, cervical, esophageal, and lung cancers, suggesting a potential role in malignant transformation and cell growth control.53 On the other hand, nucleolar 7 and PR domain zinc finger 5 (PRDM5) are proteins that are up regulated in H-genistein girls. Nuclear 7 and PRDM5 have been reported to regulate the cell cycle. The nucleolar 7 gene is reported to be a candidate tumor suppressor gene in cervical cancer that modulates the angiogenic phenotype.54 PRDM5 has growth suppressive activities and is silenced in breast, ovarian, liver, lung, colon, and other cancers.55 All four proteins should be considered as biomarkers of susceptibility for genistein/soy and cancer prevention. Interestingly, from PANTHER analysis of biological functions, the H-genistein group had the highest response on apoptotic process, a finding that corresponds very well with our report of apoptosis being increased in mammary glands of rats exposed ly to genistein.40
H-MEHP group
From the blood of girls with high MEHP concentrations in the urine, only two proteins were associated with cancer, up regulated E3 ubiquitin-ligase and insulin growth factor binding protein-3 (IGFBP-3) (Table 4). As noted earlier, E3 ubiquitin-ligase which was also up regulated in H-BPA girls is expressed in human cancer.36 On the other hand, IGFBP-3 plays a role in DNA damage repair and activation of caspase-dependent apoptosis. IGFBP-3 is a potent tumor suppressor in a variety of cancers, including breast, ovarian and prostate.56,57 Down regulated IGFBP-3 suggests less potential to repair DNA damage and to suppress cancer, and consequently it would be regarded as an event that could lead to cancer development. PANTHER biological functions also showed the highest response in the category of reproduction, a finding consistent with previous reports for animal studies with the parent compound, DEHP.18,19,58 Also, canonical pathway analysis listed five genes/proteins being differentially regulated in the category of nucleic acid metabolism and small molecule biochemistry i.e., ATP-binding cassette, chromodomain-helicase-DNA-binding 8,DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 1,huntingtin and myosin-3.
H-MBzP group
From the 47 proteins differentially regulated in the blood of girls with high urine concentrations of MBzP, three have strong cancer associations. Krueppel-like factor-10 (KLF10), RAD54B and retinoblastoma-associated protein (RB1) are all up regulated (Table 5). The KLF10 protein has been shown to inhibit the growth of cancers, and the loss of KLF10 expression in advanced pancreatic cancer is correlated with altered methylation status.59 The RAD54B gene is highly expressed in testis and spleen, which suggests active roles in meiotic and mitotic recombination. Homozygous mutations of this gene were observed in primary lymphoma and colon cancer.60 This gene is involved in homologous recombination and repair of DNA. RAD54B protein binds to double-stranded DNA and displays ATPase activity in the presence of DNA. RB1 is a negative regulator of the cell cycle and acts as a tumor suppressor. The function of phosphorylated RB1 is to prevent excessive cell growth by inhibiting cell cycle progression until a cell is ready to divide. Defects in this gene are a cause of the childhood cancer retinoblastoma, bladder cancer, and osteogenic sarcoma.61 Up regulation of these three proteins in the H-MBzP group argues for tumor suppression. However, cancer prevention has not yet been demonstrated for BBP/MBzP. On the other hand, IPA canonical pathways of function identified several proteins in H-MBzP girls that are associated with cellular assembly and organization, cellular function and maintenance: up regulated apolipo E, citron Rho-interacting kinase, granzyme B, megakaryocyte-associated tyrosine-kinase, optic atrophy 1, RB1, serum paraoxonase/arylesterase1, TAOK2 and von Willebrand factor, and down regulated fibrocystin and Ras association domain-containing 1. As seen here, proteomic data should find use in identifying biomarkers of susceptibility of other diseases.
Validation of differentially regulated proteins by western blot analysis
The use of specific antibodies provides an independent method of protein identification and quantification from that of mass spectrometry. For this, we used Western blot analysis of mammary glands of rats exposed lactationally to BPA or genistein to investigate changes in the expression levels of DLC-1, ECE-1 and EIF3. These proteins were selected based on their potential impact to predispose for or prevent cancer, and the availability of commercially prepared antibodies. Consistent with the results obtained with TMT-MS, Western blot analysis showed that in mammary glands of 50day old female rats whose dams were gavaged with BPA from days 2-20 postpartum, DLC-1 was down regulated (50% decrease) (Figure 2). Also, from mammary glands of 50day old female rats lactationally exposed to genistein during the period, ECE-1 and EIF3 were down regulated (60% and 51%, respectively).
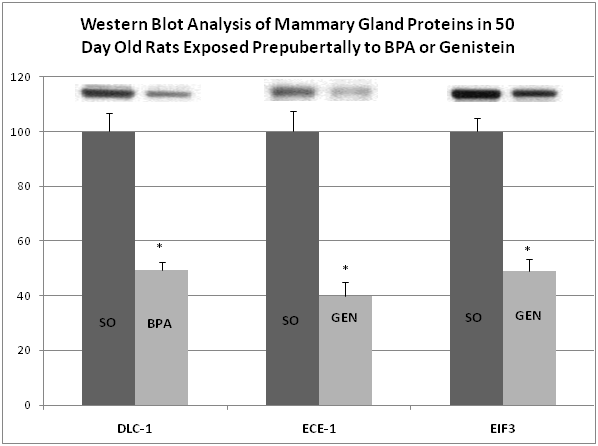
We are aware of limitations of this report, including sample size and unknown origin of the proteins assessed in the blood i.e., breast, liver, kidney, blood cells, etc. Likewise for proteins that have multiple target organ/tissue origins, we do not know the individual environmental (endogenous or exogenous) regulations. One surprising observation is the lack of common protein biomarkers in rat sera32 with human blood from the same chemical exposure groups. One difference in the methods for rat and human blood enrichment is the use of rat vs. human antibodies for protein enrichment. Other differences may be due to species metabolism, regulation, or exposures (controlled one-chemical exposure in rats versus multiple chemicals in humans), and/or a combined number of regulating caveats between the two species. On the other hand, we demonstrate in this report that the TMT-MS technology is reliable as evidenced by cross-species and tissue validations of three proteins from the blood of the girls and the mammary glands of rats exposed to BPA and genistein using Western blot analysis. This is consistent with our work on biomarkers from blood of rats exposed to BPA and genistein where we demonstrated by Western blots from blood sera similar outcome as recorded from TMT-MS sera expression.32 Also, we have recently reported that exposure of rats to BPA resulted in up regulated Ki67 protein expression in mammary glands,40 in effect validating another protein shown by TMT-MS to be up regulated in the H-BPA girls.
Via the use of TMT-MS, we demonstrate unique protein signatures that can serve as biomarkers of effect from the blood of girls in the top quintiles for urine concentration of the following environmental chemicals, BPA, MEHP, MBzP and genistein. Using bioinformatics and focusing on cancer as a disease, we also identified cancer biomarkers of susceptibility for BPA and genistein exposures. The differentially regulated cancer associated proteins in H-BPA and H-genistein girls are especially convincing in light of divergent functions and the literature demonstrating that BPA and genistein exposures are associated with mammary cancer causation and prevention, respectively.11–13,31,32,62–64 Functional identification of proteins from blood of girls with urine high concentrations of MEHP and MBzP suggests that DEHP/MEHP may contribute to carcinogenesis, while BBP/MBzP may suppress cancer development. For the latter two sets of chemicals, the potential for carcinogenesis needs to be determined. Altogether, this data demonstrates the potential of proteomic technology to not only provide biomarkers of effect from aminimally invasive source of biological material, blood, but to identify protein molecules that are intimately involved in the pathobiology of cancer. Studies with a larger cohort and long term observational studies are recommended. It is hoped that identifying early functional biomarkers of cancer will lead to intervention at the preclinical stage where susceptibility may be addressed through limiting or maximizing further exposure, as well as reversing or promoting the process, depending on whether cancer promotion or protection is afforded by the exposure.
This research was supported by Genes, Environment and Health Initiative grant 1U01ES016003, the Breast Cancer and the Environment Research Program grants U01ES/CA019482, U01ES012771, U01ES019453 and U01ES019457 from the National Institute of Environmental Health Sciences (NIEHS) and the National Cancer Institute (NCI), the UAB Comprehensive Cancer Center grant NCI 5P30CA013148, P30ES006096 from NIEHS; and UL1RR026314 from the National Center for Research Resources (NCRR).The processing and analysis of the serum samples via mass spectrometry were carried out by Drs. James Mobley and Kyoko Kojima from UAB’s Department of Surgery, Division of Urology and Comprehensive Cancer Center. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS, NCI or NIH.
The author declares no conflict of interest.

©2015 Wang, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.