
Research Article Volume 4 Issue 3
A survey on recurrent neural network based modelling of gene regulatory network
Sudip Mandal,1
Regret for the inconvenience: we are taking measures to prevent fraudulent form submissions by extractors and page crawlers. Please type the correct Captcha word to see email ID.

Goutam Saha,2 Rajat K Pal3
1Department of Electronics and Communication Engineering, Global Institute of Management and Technology, India
2Department of Information Technology, North-Eastern Hill University, India
3Department of Computer Science and Engineering, University of Calcutta, India
Correspondence: Sudip Mandal, Department of Electronics and Communication Engineering, Global Institute of Management and Technology, Krishna Nagar, West Bengal, India, Tel +919933320422
Received: October 15, 2016 | Published: November 11, 2016
Citation: Mandal S, Saha G, Pal RK. A survey on recurrent neural network based modelling of gene regulatory network. MOJ Proteomics Bioinform. 2016;4(3):244-254.
DOI: 10.15406/mojpb.2016.04.00125
Download PDF
Abstrat
The correct inference of gene regulatory networks (GRN) remains as a fascinating task for researchers to understand the detailed process of complex biological regulations and functions. With availability of large dimensional microarray data, relationships among thousands of genes can be extracted simultaneously that is a reverse engineering problem. Among the different popular models to infer GRN, Recurrent Neural Networks (RNN) are considered as most popular and promising mathematical tool to model the dynamics of, as well as to infer the correct dependencies among genes from, biological data like time series microarray. RNN is closed loop Neural Network with a delay feedback. By observing the weights of RNN model, it is possible to extract the regulations among genes. Several metaheuristics or optimization techniques were already proposed to search the optimal value of RNN model parameters. In this review, we illustrate different problems regarding reverse engineering of GRN and how different proposed models can overcome these problems. It is observed that finding out the most suitable and efficient optimization techniques for the accurate inference of small artificial, large artificial, Dream4 Network, and real world GRNs with less computational complexity are still an open research problem to all.
Keywords: gene regulatory network, recurrent neural network, microarray data, metaheuristics, optimization, regularization, cardinality, decoupling
Abbreviations
GRN, gene regulatory network; RNN, recurrent neural network; BA, bat algorithm, E. Coli, escherichia coli; SOS, save our ship; NFL, no free lunch; PSO, particle swarm optimization; ACO, ant colony optimization; GA, genetic algorithm; DE, differential evolution; ABC, artificial bee colony; IWO, invasive weed optimization
Introduction
Genes act as blue print of every living object’s activity. A gene synthesizes or produces different protein(s). A protein or a set of proteins produced by any gene, help to switch on or off the protein formation activity of other genes.1–3 Therefore, a gene can regulate other genes activities. Gene regulation is a general name for a number of sequential processes, the most well known and understood being transcription and translation, which control the level of a gene’s expression, and ultimately result with specific quantity of a target protein. A GRN is a collection of DNA segments of chromosome in a cell which interact with each other indirectly (through their protein products) and with other substances in the cell, thereby governing the expression levels of mRNA and proteins. GRN is unique and different for particular function of cells or body. As an example, different sets of genes are activated or regulated in unique way for brain cancer and blood cancer. So, GRNs for these two types of cancer will be different but unique in nature. So, inferences of these unique GRNs are very important task for researcher as it help to understand the complex phenomenon occurs inside the cells during different activity and also may help to design drugs to cure particular diseases in future.
From computational point of view, a gene regulatory network is represented by a model or graph which represents regulations or interactions amongst genes using a directed graph. In gene networks, nodes represent genes and edges represent relations or interaction amongst genes (e.g., activation or suppression) i.e. gene regulations. The regulatory relationships (depending on the nature of the control) may be of two types, namely,
- Activation: where expression value of the target gene increases, and
- Suppression: where the expression value of the target gene decreases.
Thus, a GRN can be represented by a weight matrix 𝑊=[𝑤]Nx𝑁 where the number of genes in the GRN is equal to 𝑁.If there is no regulation between two genes, value of corresponding weight is zero and if there is a regulation, the weight should have finite value. Figure 1 shows an example of GRN.

Figure 1 An example of GRN, where arrow-head and T-head denote activation and suppression respectively.
Thus, genes form a gene regulatory network, study of which appears to be very important to find the cause of a disease and the solution thereof i.e., ‘Drug design’ which is a sort of reverse engineering activity. However, GRN can be reconstructed in two ways. In Wet Lab, scientist and biologist directly identify the regulations in laboratory/ research centers. On the other hand, in case of Dry Lab, the computer science researchers reconstruct the GRN indirectly by analyzing the biological databases which are readily available online4–6 known as microarray data.
A DNA microarray7,8 (also commonly known as DNA chip or biochip) is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels9,10 of large numbers of genes simultaneously or to genotype multiple regions of a genome. Each DNA spot contains picomoles (10−12 moles) of a specific DNA sequence, known as probes (or reporters or oligos). Probe-target hybridization is usually detected and quantified by detection of fluorophore-, silver-, or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target.11,12
Gene expression levels can be determined for samples taken: 1) dynamic or time series microarray: at multiple time instants of a biological process (different phases of cell division) or 2) static or classification based microarray: under various conditions (tumor samples with different histopathological diagnosis). Each sample corresponds to a high-dimensional row vector of its gene expression profile.
Static microarray consists of gene expression to different genes for different samples those are may be in different states. Each row corresponds to gene expression values and column corresponds to different sample or patient. These type of microarray normally used for classification or prediction of disease. In case of time series microarray, each row corresponds to gene expression values and column corresponds to different time instances. However, current progresses in time series DNA microarray technologies have helped biologists a lot to examine the dynamic behaviors of different genes and the regulations among themselves by analyzing these huge gene expression values corresponding to all genes at different time instances.
Many types of models13 have been already proposed to infer gene regulatory networks and dynamics of genes from the time series microarray data i.e. a reverse engineering problem. Modelling techniques can be categorized in two types of modelling namely logical models and continuous model. Logical Models: The most basic and simplest modeling methodology is discrete and logic-based model. Logical models represent the local state of each entity in the system (such as, genes, proteins) at any time as a discrete level, and the temporal development of the system is often assumed to occur in synchronous, discrete time steps. Discrete modeling allows researchers to rely on purely qualitative knowledge. Logical models require discretization of the real valued data, which reduces the accuracy of the data. Boolean and Bayesian Network are most popular logical models. Boolean networks14,15 examine binary state transition matrices to search patterns in gene expression depending on a binary state function. Boolean Networks are ultimately limited by their definition: they are Boolean and synchronous. In reality, of course, the levels of gene expression do not have only two states but can assume virtually continuous values. Boolean is not suitable for prediction of dynamics of gene expression data, they can only show the probabilistic dependency among genes. A Bayesian network16,17 makes conditional probabilistic transitions between network states that merge the features of Hidden Markov model to include the feedback. Although effective in dealing with noise, incompleteness and stochastic aspects of gene regulation, Bayesian Network fail to consider temporal dynamic aspects that are an important part of regulatory networks modeling. Moreover, the types of regulation are also unknown. Continuous Model: Biological experiments usually produce real, rather than discrete valued, measurements. Continuous models, using real valued parameters over a continuous timescale, allow a straight- forward comparison of the global state and experimental data and can theoretically be more accurate. Continuous models13,18 are two types linear and nonlinear model. In case of linear model, In other words, the change in the level of each entity depends on a weighted linear sum of the levels of its regulators. This assumption allows a high level of abstraction and efficient inference of network structure and regulation functions. Neural Network19,20 was such types of methodology to infer GRN successfully. However, NN models failed to capture nonlinear dynamics aspects of genes regulation. When higher sensitivity to detail is desired, more complex models are preferable. On the other hand, nonlinear models encode a gene network as a system of differential equations. Difference and differential equations allow more detailed descriptions of network dynamics, by explicitly modeling the concentration changes of molecules over time. Recurrent Neural Network (RNN), S-System (SS) and General Rate Law of Transcription (GRLOT) have been widely used to model dynamic GRNs. One major advantage of all three methods lies in their simple homogeneous structures, as this allows the settings of parameter discovering software to be easily customized for these structures. In addition, all three modeling methods either already have the potential to describe additional levels of detail, or their structures can be easily extended for this purpose.
RNN21 which is a closed loop Neural Network with a delayed feedback variable, and it is suitable to model genetic system dynamics from temporal data. S-system22–26 is also another popular model based on Biochemical System Theory, which represents a GRN as a set of differential equations with power law function. When modeling GRNs with the S-system method, the expression rates are described by the difference of two products of power-law functions, where the first represents the activation term and the second the degradation term. GRLOT models27 multiply individual regulatory inputs. Activation and inhibition are represented by different functional expressions that are similar to Hill kinetics, which allow the inclusion of cooperative binding events. The degradation of gene products is defined via mass-action kinetics. Following table shows the comparative studies between these three continuous models.
Therefore, due to its less computational complexity, simplicity in network structure prediction and capability of dynamics prediction; RNN is preferred over others continuous models. Generally, RNN along with an optimization method is used to infer the GRN using microarray as training data and training error as the objective function of optimization. In this paper, we review the concept RNN and observe some existing method for modelling of GRN based on RNN that may help to beginners who are interested to carry out their research in the field of computational biology and bioinformatics. Moreover, we also emphasis some major issues and their solutions during optimization of RNN model for GRN. Next, some open research problems in this domain are also explored followed by conclusion section and references.
Theoretical background
Preliminary of RNN based GRN
As the inputs of a classical Artificial Neural Networks are supplied only from the training data of present time instance, NNs are not suitable to model the dynamics of a system. However, RNN model is a closed loop NN with a delay variable between the outputs of each neuron in the output layer of the RNN to each of the neurons in the input layer that is appropriate to model temporal data. Moreover, the regulatory relationships among a group of genes can also be expressed with the help of the canonical Recurrent Neural Network formalism28 as shown in Figure 2. Each node symbolizes a particular gene and the edges between the nodes represent the regulatory interactions among the genes.
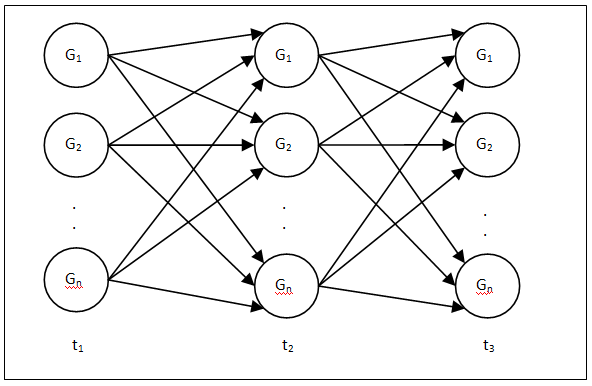
Figure 2 RNN model description of a gene regulatory network for t=t1 to t=t3.
Each tier of the RNN defines the genetic expression level of the genes at a specified time
. The gene expression level of i-th gene at a time
can be derived from the genetic expression level
of all the genes at the preceding time
, and the corresponding weights (Wi,j) between others gene with that particular gene. For a canonical RNN model, it is assumed that each of the total N output neurons in the unit is a gene expression value of next time instance ei (t+∆t), and the input neurons are the gene expression of present state ei(t) for same genes, thus they interact with each and every one in regenerative way.
(1)
Here,
is usually a sigmoid function
which is used as a classification function;
is the weight between two nodes, and it stands for the type and strength of the regulatory interaction of the
-th gene with the i-th gene. The term
represents the basal expression level or a bias term, and
is a time constant (delay) of the i-th gene;
is incremental time instance, normally it is set as 1. Thus, any RNN model which is shown in Figure 3, can be expressed by a set of parameters,
where
.
Estimation criterion
The RNN formalism is based on a set of parameters, wi,j,βi,τi which we refer to as RNN model parameters in this paper. The reverse engineering of GRNs from temporal microarray data requires finding the optimum values of the RNN parameters with the help of suitable optimization techniques or metaheuristic. However, the principle underlying all optimization algorithms is the same. Every algorithm is initialized with a population of solutions i.e. different set of
and the solutions are updated according to its optimization rules such that the training error is minimized during iteration. The time series microarrays are used for learning the optimal values of RNN model parameters.

Figure 3 A neuron in the RNN model.
wi,j is the most significant term for a GRN as its value is the connecting weight of an edge between gene-i and gene-j of the GRN,. Following are the rules for reconstruction of GRN using RNN model.
- A positive value of wi,j represents activation of gene-i by gene-j, [edge exist]
- A negative value of wi,j denotes repression or inhibition of gene- i by gene-j, [edge exist]
- A zero value means gene-j has no regulatory control on gene-i. [no edge]
All optimization methods use an objective function or a fitness function to measure the goodness of a solution. Most common estimation criterion for GRN inference from time series data is mean squared error which is defined as follows
(2)
where N is the number of genes in the problem, T is the number of time instances of the observed gene expression data and M is number of training dataset.
is numerically calculated gene expression value of k-th dataset at t-time of i-th gene using the training data from previous sampling point and RNN model parameters.
Is the actual gene expression level of k-th dataset at t-time of i-th gene. The
denotes total mean squared error between the calculated and the observed gene expression data. Therefore, RNN modeling is a non-linear function optimization problem to discover the optimal RNN parameters by minimizing the fitness function or mean square error so that calculated gene expression data fits with the observed gene expression data.
Few major issues regarding optimization for RNN parameters
During optimization of RNN based GRN, some difficulties arise which are described as follows.
Major Issue 1- Computational Complexity: Since, for N genes,
parameters must be determined to find the solution of a set of equations as in (1), a metaheuristic finds optimal RNN model parameters in
dimensional search space. However, this space becomes very computationally expensive in case of large-scale genetic networks. Therefore, optimization of this huge number of parameters in a single program is quite difficult which may lead to erroneous results and large computation time. Complexity is needed to be reduced so that runtime is minimal and inference accuracy can increase also. Moreover, algorithm should able to infer GRN from less number of time series data without stuck at local optimal point.
Major Issue 2- accuracy in prediction of dynamics of genes: Learning the RNN model parameters so as to fit best the predicted expression dynamics with the training data is, in essence, an optimization problem. Normally in RNN based Gene Regulatory network reconstruction using different metaheuristic, two criterions are needed to be satisfied. First one is the minimization training error
as in Eqn. (2), which leads us to correct prediction of dynamics for the gene expressions.
Moreover, the inferred values of the regulatory parameters are also bit of concern as its magnitude can affect the network connectivity. So, we define another performance measurement parameter as Inferred Parametric Error (IPE) which measures the deviation in magnitude of inferred parameters of RNN model from original one.
(3)
Where
are the actual values of RNN parameter and
are the calculated value of the same. Using these two types of performance parameter, we can estimate the efficiency of an inference algorithm. Both training error and IPE should be small as much as possible for accurate prediction of dynamics of genes.
Major issue 3- over fitting problem:Another most important criterion, which should be kept in mind, is the structure of the gene regulatory network i.e. correct regulations in the network. Real-life genetic networks are sparsely connected29 i.e. very few connections exist between the genes. It may be possible that though the dynamics is correctly predicted i.e. corresponding training error is very small, but the network structure is completely different (i.e. different set of
for RNN) due to different optimal solutions (i.e. local optima) found by the metaheuristic and which is known as over-fitting problem. Moreover for this problem, true regulations are missing and many false regulations are included in the reconstructed network.
Now, the performance of an algorithm for correct network prediction is measured in terms of its sensitivity
, specificity
, accuracy (A) and F-score
which are defined as follows
(4)
(5)
(6)
(7)
TP (True Positive) denotes the number of correctly predicted regulations, and TN (True Negative) represents the number of properly predicted non-regulations. FP (False Positive) denotes the number of incorrectly predicted regulations, and FN (False Negative) represents the number of falsely predicted non-regulations by the inference algorithm. Sensitivity denotes the fraction of the total number of existing edges in the original network, correctly predicted in the inferred network. Specificity denotes the fraction of the total number of non-existent edges in the original network, correctly identified as non-existent in the inferred networkas well. Accuracy denotes the capability of inferencing all true existing and nonexisting regulations.F-score is calculated to evaluate an algorithm without looking at the trade-off between sensitivity and specificity. The values of sensitivity
, specificity
, accuracy (A) and F-score
should be ideally 1 for accurate inference of GRN.
Solutions addressed regarding above issues
To overcome these problems during optimization following actions may be taken.
Decoupling to reduce computational complexity: To overcome computational complexity problem, the genetic network inference problem can be divided or decoupled30 into several sub-problems for a single gene. The change in expression level of a particular gene at a given time instant depends on the expression levels of all genes at previous time instant only. Moreover, the changes in expression level for different genes in that given time instant are independent of each other. Therefore, a decoupling procedure can be introduced here without losing any vital information. Now, the objective function
for this decoupled RNN31 is the training error for the i-the gene only
(8)
Hence, the numbers of RNN parameters are needed to determine is only
parameters for i-th gene. Thus, this decoupling method divides an N(N+2)-dimensional problem space into
-dimensional sub problem space for each gene. Instead of finding
parameters in single program run, decouple method focus to optimize
parameters in a single run and execute the program for
time for
different genes. By accumulating the
parameters of all N genes, overall structure of RNN can be achieved which in turn denotes the GRN. Moreover less number of training data must be used so that computational time of the algorithm.
Selection of suitable optimization technique to increase accuracy:Moreover, No Free Lunch (NFL) theorem31 logically states that there is no single metaheuristic which is best suited for solving all kind of optimization problems. Therefore, new and suitable metaheuristic must be employed for accurate prediction in dynamics of gene expression so that training error and IPE are minimized. Moreover, it is possible to modify and tune the existing metaheuristic for faster convergence and better accuracy.
Regularization to deal with over-fitting problem:To overcome over-fitting problem, the researchers normally add different regularizer to the training error which acts as objective function of the optimization process to balance between actual network structure and dynamics of genes. Regularizer normally acts penalty term that adds penalty term for different architecture of the GRN. In literature, many kind of penalty term were proposed, which will be discussed in next section.
How to validate a new algorithm for RNN based GRN reconstruction?
For RNN modelling of GRN, the performances of a new algorithm must be validated against following artificial and real life genetic networks (benchmark problems) to prove its efficiency.
- 4 Genes Artificial network without and with noise
- 30 Genes Artificial network without and with noise
- Real world Genetic Network like E. Coli 32
- Synthetic Network like Gene Net Weaver Network33
Following Table 1, 2 & Table 3 show the parameters of 4 gene artificial network and 30 gene artificial network respectively which are benchmark problems in this domain of research. Several authors34-38 used these parameters to generate artificial time series data and infer artificial network for validation using these generated dataset. Authors38 also add different level Gaussian noise to the data and check robustness of their proposed algorithm in presence of noise.
Characteristics |
RNN |
S-System |
GRLOT |
Number of parameters to be optimized |
N*(N+2) |
N(2N+2) |
N(2N+1) |
Limitation (saturation) |
Yes |
No |
Yes |
Processing of gene expression |
OR |
AND |
AND |
Regulation of Degradation processes |
No |
Yes |
No |
Regulations and its types depends on |
Only on weights (w) |
both kinetic parameter (gij and hij) |
Hill exponent nij , Kak , Kij |
Computational Complexity |
Moderate |
Large |
Large |
Inference of regulation |
Simple |
Complex |
Complex |
Table 1 Comparative studies between RNN, SS, and GRLOT
wi,j |
1 |
2 |
3 |
4 |
βi |
τi |
1 |
20 |
-20 |
0 |
0 |
0 |
10 |
2 |
15 |
-10 |
0 |
0 |
-5 |
5 |
3 |
0 |
-8 |
12 |
0 |
0 |
5 |
4 |
0 |
0 |
8 |
-12 |
0 |
5 |
Table 2 RNN Model Parameters for Small Artificial Network
SD: Standard Deviation; BMI: Body Mass Index; WC: Waist Circumference; AC: Abdominal Circumference; HC: Hip Circumference; RER: Respiratory Exchange Ratio; HR: Hear Rate.
|
|
|
|
,
|
|
|
|
Table 3 RNN Model Parameters of Large Artificial Network
The Save Our Ship (SOS) network for E. Coli 29 was first introduced by Uri Alon group39 which is a benchmark in GRN problem to find out the effectiveness of the inference algorithm34,36,37,40-42 on real time dataset43 and network. In SOS network, 8 genes were considered (uvrD, lexA, umuD, recA, uvrA, uvrY, ruvA and polB). During their experiments, DNA of E.Coli was irradiate with the UV light, which affected some gene, after that, the network would repair itself by suppressing others genes expression value. They performed four experiments for different UV light intensities. Each experiment consists of 50 time steps spaced by 6 minutes for each of the eight genes (Figure 4).
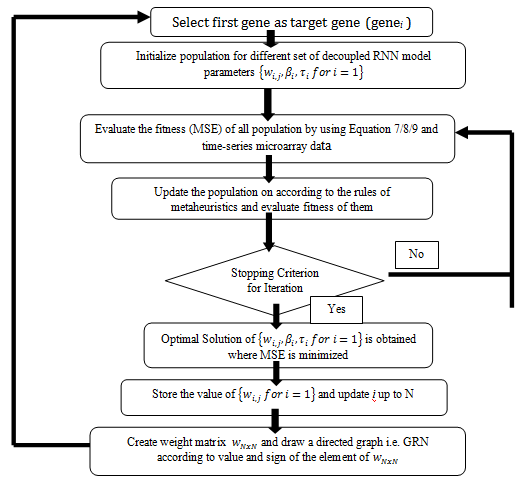
Figure 4 A typical flowchart for inference of GRN based RNN model using any met heuristic.
Since the yearly DREAM challenge keeps providing all kinds of benchmarks, inference of Synthetic Network generated by Gene Net Weaver (version 3.1.3 beta)33 is another interesting benchmark problem for researchers.
Literature survey
It has been a long history that RNN model along with different intelligent methods were widely used in bioinformatics for inference of Gene Regulatory Network. A typical flowchart for inference of GRN based on decoupled RNN model using any metaheuristic is described in Figure 4. For example, Hu X44 used Back Propagation through Time (BPTT) learning rule to learn both weight matrix and decay time constant of RNN model. The proposed approach was tested for reconstruction of E. Coli SOS network. Predicted dynamics of genes expression were deviated significantly from the actual one. Moreover, using standard deviation and variance analysis of outputs, it detected 6 true regulations but also included 11 unwanted regulations during reconstruction of GRN.
In 2007, Xu R 45 used Particle Swarm Optimization (PSO) technique to infer RNN based GRN without any regularization term. It is observed that performance of PSO greatly depend on parameters selection and only weight are optimized using PSO with 50 points time series data. For small artificial network, author mainly focused on prediction of dynamics though weight values are not correct inferred and that leads to a huge IPE for this model. Moreover, 1 FP and 1 FN are also detected. In case of E. Coli Network, nonlinear dynamics is predicted with satisfactory accuracy but able to detect only 5 TP and 2 FP (best result) at certain binomial distribution probability p.
In 2007 Xu R 45 proposed a new hybrid evolutionary–swarm algorithm DEPSO, based on the combined concepts of PSO and Differential Evolution (DE), to address the challenge of training RNNs. The performance of DEPSO is also compared with PSO and DE and the simulation results demonstrate the effectiveness of the hybridization of different evolutionary/swarm technologies in improving their search capability in network parameters learning. The author mainly focused on dynamics prediction of 8 Genes synthetic network with 30 point time series data but lots of FPs were included in the network and weight values are not correct. For E. Coli network, dynamics is correctly predicted but, only 2 out of 9 potential connections were correctly identified.
On the other hand, Chiang JH46 introduced a Genetic Algorithm Recurrent Neural Network (GARNN) hybrid method for finding feed forward regulated genes when given some transcription factors to construct cancer related regulatory modules from human cancer microarray data. This hybrid approach focused on the construction of various kinds of regulatory modules, by considering the fact that Recurrent Neural Network has the capability of controlling feed forward and feedback loops in regulatory modules and Genetic Algorithms provide the ability of global searching of common regulated genes. This approach unraveled new feed forward connections in regulatory models by modified multilayer RNN architectures. The model was validated against human cell cycle and Yeast cell cycle microarray data.
In 2008 Lee WP35 dealt with the scalability problem of RNN based GRN by developing a clustering method with several data analysis techniques. The author used BPTT learning rule for training of RNN whereas Self Organization feature Map (SOM) and Wavelet Transform (WT) for gene clustering. Here authors also only concentrated on dynamics of the model and was tested for 2,4,10 genes artificial network.
In 2009, Zhang Y47 used a hybrid of particle swarm optimization and recurrent neural network (PSO-RNN) methods with un regularized fitness function to infer the underlying network between modules. The author improves the inference accuracy of a GRN by (1) incorporating prior biological knowledge into the inference scheme, (2) integrating multiple biological data sources, and (3) decomposing the inference problem into smaller network modules. The method was validated on real data from two cases: the development of rat central nervous system (CNS) and the yeast cell cycle process.
In 2010 Maraziotis IA,48 proposed an approach based on a novel multilayer evolutionary trained neuro-fuzzy recurrent network (ENFRN) that was able to select potential regulators of target genes and describe their regulation types. The recurrent, self-organizing structure and evolutionary training of the network yield an optimized pool of regulatory relations, while its fuzzy nature helped to avoid noise-related problems. Furthermore, the assign scores for each regulation, highlighting the confidence in the retrieved relations. The approach was tested against several benchmark datasets of yeast and managed to acquire biologically validated relations among genes.
In 2011, Ghazikhani A49 proposed a multi agent system for RNN training. The agents of the proposed multi agent system trainer were separate swarms of particles building up a multi population Particle Swarm Optimization (PSO) algorithm. Multi Agent System was comprised of three types of agents, parent agent, structure PSO agent and parameter PSO agent. The model can able to infer only 2 out of 9 potential connections correctly for SOS network of E. Coli. So, its performance was very poor.
In 2012, Rakshit P36 used Hybrid Invasive Weed Optimization and Ant Bee Colony Optimization along with fuzzy based RNN for inference problem. They propose a novel cost function along penalty term
(8)
where p is the weight constant that denote magnitude of penalty to balance between over fitting and actual network structure.
be a defuzzified weight from neuron j to neuron i in the neural net. But inference process was quite complicated and there were huge parametric error in weights but dynamics is accurate for small genetic network. Moreover, it perform very poor for E. Coli SOS Network where lots of FPs were included and regulation types changed in many cases.
In 2012, Kentzoglanakis K37 used Ant Colony Optimization (ACO) and Particle Swarm Optimization for searching biologically plausible architecture where ACO was used to search the regulators of genes in discrete space, PSO search the parameters of RNN for those topology. For 4 gene artificial network, it can infer only 65% of TPs. For 10 genes GNW data, it can detect 80% TPs. For E. Coli SOS network, it can detect 8 TPs but also included 4 FPs for 2nd experiment dataset. The author also tested their algorithm against Yeast dataset. However, in all cases prediction of the dynamics is quite satisfactory but the process was very time consuming due to parallel implantation of two separate optimization technique.
In 2012, Palafox L41 implemented a classic Population Based Incremental Learning (PBIL), which in certain scenarios had outperformed classic GA and other evolutionary techniques like Particle Swarm Optimization (PSO). They tested this implementation on small and large artificial networks. The results, however, showed that the approach was adequate for large networks, and it was capable of finding results faster than ODEs approaches.
In 2013, Palafox L42,43 has used Population Based Incremental Learning (PBIL) enhanced with K-means clustering to find the optimum parameters of the Recurrent Neural Network. However, the hard clustered approach is a naive attempt to model the dynamics of the population. The algorithm was tested against only for E. Coli SOS network where 7 TPs, 3 FNs and 4 FPs are detected. The algorithm performed on par with some of the best implementations for SOS network structure. However, the hard clustered approach is a naive attempt to model the dynamics of the population.
In 2013, Noman N25 proposed a novel Decoupled Recurrent Neural Network which was trained Differential Evolution (DE) technique and introduced a penalty term i.e. L1 regularizer in the objective function to balance between minimum training error and actual network structure. The detail of this novel penalty term is given in Equation 8. Their result had shown very good accuracy in finding all true regulation and dynamics for both small and large network. The main disadvantage of this process was inclusion of large number of false regulations. In this regularization process, normally it was assumed those maximums I number of regulations are allowed by considering the fact of sparse nature of the GRN i.e. few connectives exist in real life GRN. This maximum number allowable regulation is also known as cardinality. To generate the sparse solutions for RNN, the concept of in-degree or cardinality of genes in error function was already introduced. A penalty term based maximum cardinality I was added to the fitness function for real life network reconstruction where it was assumed that out of N values of w for each of genes, only I non-zero values were allowed within each w vectors , thus forcing the other (N-I) values to become zero. This is equivalent to prior biological knowledge into the inference scheme. If any of these (N-I) elements achieved a non zero-value during optimization process, the solution will be penalized in the following way25 for decoupled RNN system
(9)
where
is the vector which contains the absolute values of
but sorted in ascending order. It performed very well for 4 genes artificial network without noise whereas 50% false positive regulations were included for 5% noise. Moreover, it identified 50% TPs and most of the TN for 30 genes artificial network but some FPs were included in noiseless case. However, its performance was very poor in presence of noise for large artificial network. For E coli SOS net, 5 TPs were detected but 11 FPs are introduced although dynamics was correctly predicted.
In 2016, Mandal S38 proposed a hybrid Cuckoo Search (CS) and Flower Pollination Algorithm (FPA), to infer large scale gene regulatory network with better accuracy. CS was used to search in discrete space to select the best combination of I regulatory genes where FPA was implemented to find out the optimal values of RNN model parameters for those regulators. Initially, the proposed method was tested on a benchmark 30 genes large-scale artificial network for both noiseless and noisy data. The results suggested that this hybrid algorithm can capable of inferring more number of correct regulations and less number of false regulations than others existing methods. Secondly, the proposed methodology has been validated against the real-world dataset of the DNA SOS repair network of Escherichia coli. However, time complexity is larger due to implementation of two optimization processes sequentially.
In 2016, Khan A50 proposed a novel Bat Algorithm-Particle Swarm Optimization (BAPSO) RNN model parameters and the results obtained show that the predicted networks reproduce the dynamics of the given dataset to a better extent for small-scale GRNs using different types of microarray datasets (synthetic, in silico, and in vivo). However, for medium-scale networks (20-gene), the performance deteriorates. There are too few true predictions and a large number of incorrect predictions. Moreover, the topology also depends on proper selection of threshold 5 which is very difficult task for unknown network.
It has been observed that all authors did not taste their algorithms for all types of genetic networks (benchmark problems) and few of them dealt only with the dynamics of gene expression data while they were not concerned about the structure of the inferred network. Moreover, few authors used different networks for their validation (for example Xu R34 used an 8 gene small artificial network instead of 4 gene benchmark network). On the other hands, some of them did not mention about true and false regulations. Therefore, comparison between all states of art techniques is slightly difficult. So, the outlines of their results in terms of Sn, Sp, A, and F are given serially in Table 4. The networks which were not validated by a particular author were written as “Not Reported” during comparison.
Authors |
Network Type |
Small Artificial |
Medium/Large Scale |
GNW Network |
Real World Network |
Without Noise |
With Noise |
Without Noise |
With Noise |
X Hu40 |
Not Reported |
Not Reported |
Not Reported |
Not Reported |
Not Reported |
Dynamics for E. Coli Network was correctly predicted.
Sn,=0.67,Sp=0.89, A=0.86, F=0.57 |
Rui Xu45 |
For 8 gene Network
Sn,=0.67,Sp=0.83, A=0.76, F=0.70 |
Not Reported |
Not Reported |
Not Reported |
Not Reported |
Dynamics of SOS network is was correctly predicted.
TP=2 but
FP, FN,TN were not reported |
Rui Xu45 |
For 4 gene network
Sn,=1,Sp=1, A=1, F=1. |
Not Reported |
Not Reported |
Not Reported |
Not Reported |
Dynamics of SOS network was correctly predicted.
Sn,=0.56,Sp=0.96, A=0.90, F=0.62 |
Chiang JH46 |
Not Reported |
Not Reported |
Not Reported |
Not Reported |
Not Reported |
Validated against human cell cycle and Yeast cell cycle microarray data. TP, FP, FN,TN were not reported |
Lee WP35 |
Focus on mainly dynamics for 2,4,10 genes artificial network |
Not Reported |
Not Reported |
Not Reported |
Not Reported |
Not Reported |
Yuji Zhang47 |
Not Reported |
Not Reported |
Not Reported |
Not Reported |
Not Reported |
The method was tested against rat central nervous system (CNS) and the yeast cell cycle
TP, FP, FN,TN were not reported |
Maraziotis IA38 |
Not Reported |
Not Reported |
Not Reported |
Not Reported |
Not Reported |
Tested against datasets of yeast cycle network
TP=11, FN=3.
FP,TN were not reported |
Ghazikhani A49 |
Not Reported |
Not Reported |
Not Reported |
Not Reported |
Not Reported |
Dynamics of SOS network were correctly predicted.
TP=2 but
FP, FN,TN were not reported |
Rakshit P36 |
For 4 gene network,
Sn,=1,Sp=0.62, A=0.81, F=0.84. |
Not Reported |
Not Reported |
Not Reported |
Not Reported |
Dynamics SOS network is predicted correctly.
Sn,=0.89,Sp=0.14, A=0.25, F=0.25. |
Kentzoglanakis K37 |
For 4 gene network,
Sn,=0.62,Sp=0.87, A=0.75, F=0.71. |
Not Reported |
Not Reported |
Not Reported |
For 10 genes GNW data, Sn=0.58, Sp=0.24. A and F were not reported |
For SOS network,
Sn,=0.89,Sp=0.90, A=0.90, F=0.72. |
Palafox L41 |
Mainly focused on 6 gene network for dynamic analysis |
Not Reported |
Not Reported |
Not Reported |
Not Reported |
Not Reported |
Palafox L42 |
Not Reported |
Not Reported |
Not Reported |
Not Reported |
Not Reported |
For SOS network,
Sn,=0.70,Sp=0.76, A=0.74, F=0.67. |
Noman N25 |
Perform very well for 4 gene artificial network.
Sn,=1,Sp=1, A=1, F=1. |
In presence of 5% noise
Sn,=1,Sp=0.50, A=0.75, F=0.80. |
For 30 gene network
Sn,=0.61,Sp=0.99, A=0.98, F=0.72. |
For 5% noise
Sn,=0.30,Sp=0.98, A=0.95, F=0.35. |
Not Reported |
Dynamics SOS network is predicted correctly.
Sn,=0.78,Sp=0.81, A=0.81, F=0.54. |
Mandal S38 |
Not Reported |
Not Reported |
For 30 gene network,
Sn,=0.89,Sp=0.99, A=0.99, F=0.88 |
For 5% noise,
Sn,=0.89,Sp=0.98, A=0.97, F=0.73. |
Not Reported |
For Ecoli SOS net,
Sn,=0.44,Sp=0.80, A=0.75, F=0.33. |
Khan A50 |
For 4 gene artificial network,
Sn,=0.75,Sp=1, A=0.87, F=0.86 |
Not Reported |
Not Reported |
Not Reported |
For 10 genes GNW, Sn,=0.50,Sp=0.86, A=0.82, F=0.40 |
For Ecoli SOS net,
Sn,=0.78,Sp=0.84, A=0.83, F=0.56. |
Table 4 Summary of Existing Work on RNN Based Inference of GRN
An algorithm should accurately infer the actual structural as well as the dynamics of gene expression data with less computational complexity. However, Differential Evolution (DE) technique is performed better than others technique for all types of networks. CS-FPA hybrid performs better only in case large artificial network in presence of noise. BAPSO is also a good choice for inferring all kinds of small scale genetic network.
Open research issue
Different metaheuristic are applied successfully to infer small scale gene regulatory network structure as well as dynamics of genes in presence of noise and without noise but yet to accomplish an accurate inference of large-scale artificial, Dream4 and real life GRNs with less computational time. However, few of them were able to found all true regulations but they also detected some false regulations. Therefore followings are still open research problems to the computer researchers in the field of RNN modeling of GRN
- Finding out the most suitable and efficient optimization techniques for the accurate inference of small artificial (without and with noise), large artificial (without and with noise), GNW or Dream4, and real world GRNs with less computational complexity.
- Modification or improvement of existing metaheuristic for the accurate inference of all kind of GRNs with less time series data. However, it is worth to mention that outputs of metaheuristics also depend on initialization (initial input data for learning) of algorithm. So, algorithms must have capability to overcome this problem.
- Modification of regularization or penalty term can be done term to obtain better result (like dynamic penalty where penalty value change with iteration to balance more etc.).
- Some techniques must be employed for reduction in FPs from the GRN.
- Hybridization of different optimization techniques may be introduced to reduce the search space of optimization (as in case of regularized cost function, the algorithm still search in large space for all parameters) by fixing the regulators size.
Conclusion
So, accurate inferences of all types of GRNs with less computational complexity, less data point but with high accuracy are still fascinating task to computer science researchers. In this paper, we have given a brief idea about inference problem of GRN i.e. reverse engineering of genetic network. Recurrent Neural Network is one of the most popular artificial techniques for GRN modeling. In this literature survey, different states of art techniques for this purpose are described where it is found that most of the authors used a metaheuristic for optimization of parameters of RNNs. Most of the techniques performed very well for small scale genetic network. Although exact inference of real world and synthetic large scale GRN are still difficult tasks to be achieved. However, dynamics of gene expression prediction is comparatively easy task than network inference. We also investigated some major issues that occurred during optimization of RNN model and also proposed some solutions regarding them. Moreover, we review the applications of different optimization techniques and hybrid intelligence methods in bioinformatics, which may help researchers or beginners from both areas to understand each other and ensure their future collaborations.
Acknowledgements
Conflict of interest
The author declares no conflict of interest.
References
- Fankhauser N, Cima I, Wild P, et al. Identification of a Gene Expression Signature Common to Distinct Cancer Pathways. Cancer Inform. 2012;11:139–146.
- Cho RJ, Campbell MJ, Winzeler EA, et al. A Genome–Wide Transcriptional Analysis of the Mitotic Cell Cycle. Mol Cell. 1998;2(1):65–73.
- Chu S, DeRisi J, Eisen M, et al. The transcriptional program of sporulation in budding yeast. Science. 1998;282(5389):699–705.
- http://smd.stanford.edu
- Gene Expression Omnibus (GEO).
- http://www.godatabase.org/cgi–bin/go.cgi
- Masys DR. Linking microarray data to the literature. Nat Genet. 2001;28(1):9–10.
- Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32 Suppl:496–501.
- Brazma A, Vilo J. Minireview: Gene expression data analysis. Federation of European Biochemical societies, 2000. 480:17–24.
- Schena M, Shalon D, Davis RW, et al. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470.
- Lockhart DJ, Dong H, Byrne MC, et al. Expression monitoring by hybridization to high–density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680.
- Shyamsundar R, Kim YH, Higgins JP, et al. A DNA microarray survey of gene expression in normal human tissues. Genome Biology. 2005;6:R22.
- Vijesh N, Kumar S, Sreekumar J. Modeling of gene regulatory networks: A review J. Biomedical Science and Engineering. 2013;6:223–231.
- Akutsu T, Miyano S, Kuhara S. Identification of Genetic Networks from a Small Number of Gene Expression Patterns under the Boolean Network Model. Pac Symp Biocomput. 1999:17–28.
- Weaver DC, Workman CT, Stormo GD. Modeling Regulatory Networks with Weight Matrices. Pac Symp Biocomput. 1999:112–123.
- Perrin BE, Ralaivola L, Mazurie A, et al. Gene networks inference using dynamic Bayesian networks. Bioinformatics. 2003;19 Suppl 2:ii138–ii148.
- Werhli AV, Grzegorczyk M, Husmeier D. Comparative evaluation of reverse engineering gene regulatory networks with relevance networks, graphical Gaussian models and Bayesian networks. Bioinformatics. 2006;22(20):2523–2531.
- Swain MT, Mandel JJ, Dubitzky W. Comparative study of three commonly used continuous deterministic methods for modelling gene regulation networks. BMC Bioinformatics. 2010;11:459.
- Keedwell E, Narayanan A. Discovering Gene Networks with a Neural–Genetic Hybrid. IEEE/ACM Trans Comput Biol Bioinform. 2005;2(3):231–242.
- Wahde M, Hertz J. Modeling Genetic Regulatory Dynamics in Neural Development. J Comput Biol. 2001;8(4):429–442.
- Kolen J, Kremer S. A Field Guide to Dynamical Recurrent Networks. IEEE Press; 2001.
- Mandal S, Khan A, Saha G, et al. Reverse Engineering of Gene Regulatory Networks Based on S–Systems and Bat Algorithm. J Bioinform Comput Biol. 2016;14(3):1650010.
- Liu LZ, Wu FX, Zhang WJ. Inference of Biological S–System Using the Separable Estimation Method and the Genetic Algorithm. IEEE/ACM Trans Comput Biol Bioinform. 2012;9(4):955–965.
- Chowdhury AR, Chetty M, Vinh NX. Incorporating Time–Delays in S–System Model for Reverse Engineering Genetic Networks. BMC Bioinformatics. 2013;14:196.
- Palafox L, Noman N, Iba H. Reverse Engineering of Gene Regulatory Networks Using Dissipative Particle Swarm Optimization. IEEE Transactions on Evolutionary Computation. 2013;17(4):577–587.
- Mandal S, Saha G, Pal KR. S–System Based Gene Regulatory Network Reconstruction Using Firefly Algorithm. Third International Conference on Computer, Communication, Control and Information Technology (C3IT–2015); 2015:1–5.
- Mendes P, Sha W, Ye K. Artificial gene networks for objective comparison of analysis algorithms. Bioinformatics. 2003;19(90002):122–129.
- Weaver DC, Workman CT, Stormo GD. Modeling regulatory networks with weight matrices. Pac Symp Biocomput. 1999;1999:112–123:
- Thieffry D, Huerta AM, Pérez–Rueda E, et al. From specific gene regulation to genomic networks: a global analysis of transcriptional regulation in Escherichia coli. Bioassays. 1998;20(5):433–440.
- Noman N, Palafox L, Iba H. Reconstruction of Gene Regulatory Networks from Gene Expression Data Using Decoupled Recurrent Neural Network Model. Proceeding in Information and Communication Technology (PICT 6). 2013:93–103.
- Wolpert DH, William G Macready. No Free Lunch Theorems for Optimization. Evolutionary Computation.IEEE Transactions on Evolutionary Computation. 1997;1(1):67–82.
- Faith JJ, Hayete B, Thaden JT, et al. Large–scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5(1):e8.
- Schaffter T, Marbach D, Floreano D. GeneNetWeaver: in silico benchmark generation and performance profiling of network inference methods. Bioinformatics. 2011;7(16):2263–2270.
- Xu R, Wunsch Ii D, Frank R. Inference of genetic regulatory networks with recurrent neural network models using particle swarm optimization. IEEE/ACM Trans Comput Biol Bioinform. 2007;4(4):681–692.
- Wei–Po Lee, Kung–Cheng Yang. A clustering–based approach for inferring recurrent neural networks as gene regulatory networks. Neurocomputing. 2008;71(4–6):600–610.
- Rakshit P, Das P, Konar A, et al. A Recurrent Fuzzy Neural Model of a Gene Reglatory Network for Knowledge Extraction Using Invasive Wee and Artificial Bee Colony Optimization Algorithm. 1st International Conference on Recent Advances in Information Technology (RAIT); 2012.
- Kentzoglanakis K, Poole M. A Swarm Intelligence Framework for Reconstructing Gene Networks: Searchnig for Biologically Plausible Architecture. IEEE/ACM Trans Comput Biol Bioinform. 2012;9(2):358–371.
- Mandal S, Khan A, Saha G, et al. Large Scale Recurrent Neural Network Based Modeling of Gene Regulatory Network Using Cuckoo Search–Flower Pollination Algorithm. Advances in Bioinformatics. 2016;2016:1–9.
- Ronen M, Rosenberg R, Shraiman BI, et al. Assigning numbers to the arrows: parameterizing a gene regulation network by using accurate expression kinetics. Proc Natl Acad Sci U S A. 2002;99(16):10555–10560.
- Hu X, Maglia A, Wunsch D. A General Recurrent Neural Network. Conf Proc IEEE Eng Med Biol Soc. 2005;5:4735–4738.
- Palafox L, Iba H. Gene Regulatory Network Reverse Engineering using Population Based Incremental Learning and K–means. USA: GECCO’12 Companion; 2012.
- Palafox L, Iba H. Study on the Use of Evolutionary Technique for Inference in Gene Regulatory Networks. Proceeding in Information and Communication Technology (PICT 6). 2013:82–92.
- http://www.weizmann.ac.il/mcb/UriAlon/download/downloadable–data
- Hu X, Maglia A, Wunsch D. A General Recurrent Neural Network. Conf Proc IEEE Eng Med Biol Soc. 2005;5:4735–4738.
- Xu R, Venayagamoorthy GK, Wunsch DC II. Modeling of gene regulatory networks with hybrid differential evolution and particle swarm optimization. Neural Net. 2007;20(8):917–927.
- Chiang JH, Shih Yi Chao. Modeling human cancer related regulatory modules by GARNN hybrid algorithms. BMC Bioinformatics. 2007;8:91.
- Zhang Y, Xuan J, de los Reyes BG, et al. Reverse engineering module networks by PSO–RNN hybrid modeling. BMC Genomics. 2009;(Suppl 1):S15.
- Maraziotis IA, Dragomir A, Thanos D. Gene regulatory networks modelling using a dynamic evolutionary hybrid. BMC Bioinformatics. 2010;11:140.
- Ghazikhani, Akbarzadeh TMR, Monsefi R. Genetic regulatory network inference using Recurrent Neural Networks trained by a multi agent system. Iran: Computer and Knowledge Engineering (ICCKE), 2011 1st International eConference on Mashhad; 2011 p. 95–99.
- Khan A, Mandal S, Pal RK, et al. Construction of Gene Regulatory Networks Using Recurrent Neural Networks and Swarm Intelligence. Scientifica (Cairo). 2016;2016:1060843.

©2016 Mandal, et al. This is an open access article distributed under the terms of the,
which
permits unrestricted use, distribution, and build upon your work non-commercially.




