MOJ
eISSN: 2374-6920


Research Article Volume 7 Issue 3
1Bioinformatics Centre, Gurucharan College, India
2Department of Biotechnology, Gurucharan College, India
Correspondence: Manabesh Nath, Research Scholar, Bioinformatics Centre, Gurucharan College, Silchar, Assam-788004, India
Received: March 22, 2018 | Published: June 22, 2018
Citation: Nath M, Deb B. A computational approach of antibiotics as novel drug target for Japanese encephalitis virus NS helicase/nucleoside triphosphatase. MOJ Proteomics Bioinform. 2018;7(3):184 ? 189. DOI: 10.15406/mojpb.2018.07.00231
Japanese encephalitis (JE) is a mosquito-borne viral neurologic disease caused by an RNA virus–Japanese encephalitis virus (JEV) of JEV serocomplex belonging to genus Flavivirus of family Flaviviridae. For the purpose of the study, we selected few candidate compounds belonging to Aminoglycosides and Tetracycline category that are known to be protein synthesis inhibitors. JEV NS3 helicase/NTPase (PDB ID: 2Z83) protein was the receptor of interest. Drugs Kanamycin, Rolitetracycline, and Doxycycline showed better binding energy compared to two study standards- Ribavarin and Minocycline tetracycline. Interacting bonds were formed in all three domains of NS3 helicase/NTPase, particularly in motifs I and VI of helicase which is important in viral replication. Hence, we can state that on the basis of virtual screening, the antibiotics could be suggested as a possible novel therapy against JE disease which warrants further in vitro screening.
Keywords: japanese encephalitis, NS3 helicase/NTPase, docking, virtual screening,kanamycin, doxycycline
JE, japanese encephalitis; CNS, central nervous system; JEV, japanese encephalitis virus; NTP, nucleoside triphosphate
Japanese encephalitis (JE) is an arthropod-borne viral disease of global public health importance. The disease is prevalent in Southeast Asia and the Pacific.1 However, scientific evidence suggests that JE will soon become a global pathogen and cause of worldwide pandemics.2 JE causes the central nervous system (CNS) disease with irreversible neurological damage in humans.3 Annually 13,600 to 24,400 deaths due to acute JE infection are recorded. JE survivors with an annual report of 14,300 to 24,400 cases develop long-term neuropsychological sequelae.4 JE is caused by Japanese encephalitis virus (JEV) which belongs to genus Flavivirus under family Flaviviridae. JEV is transmitted to humans through the bite of infected Culex species mosquitoes.5 However, there is no effective antiviral therapy and only a few highly effective vaccines are licensed for human use. The search for antiviral products becomes very important. Virtual screening has repeatedly proven to be useful to meet the special challenges of antiviral drug discovery.
The open reading frame (ORF) of JEV genome translates into a large polyprotein containing 10 proteins. The N- terminal of one-fourth of polyprotein encodes structural proteins- Capsid (C), Membrane glycoprotein (prM) and Envelope (E) and non-structural (NS) proteins (NS1-NS2-NS2B-NS3-NS4A-NS4B-NS5). NS3 of JEV is a multifunctional protein of 619 amino acid residues. It possesses enzymatic activities of a serine protease, helicase and nucleoside 5'-triphosphatase (NTP). One-third of N terminal end of NS3 has a serine protease activity that participates in the processing together with a cofactor protein NS2B. The NS3 protein also has a catalytic domain for helicase, NTPase, as well as 5′-terminal RNA triphosphatase activities in the C-terminal two-thirds. NS3 plays important roles in the processing of the viral precursor polyprotein and replication of viral genomic RNA. NS3 serves as a reservoir for viral proteins during virus assembly.6,7 Helicase is grouped into three superfamilies and one family based on several conserved motifs.8 Motifs I is also known as Walker A which is a phosphate binding P-loop. Motif II, also known as Walker B has a Magnesium binding aspartic acid loop. Both the motifs exist in all helicase superfamilies.9 Helicase can be further classified into DEAD/H, DExD, and DExx subfamilies based on the sequence of motif II.8 The flavivirus helicase is a member of the DEAH/D box family within helicase superfamily 2.10 NS3 Helicase/ NTPase is essential for viral life cycle and this structure motif could be an ideal target for the development of novel potent therapeutic regimen.7 Yamashita T et al.,5 examined three- dimensional structure of JEV helicase/NTPase domain (amino acid residues 171-619). JEV NS3 helicase comprises of three domains. Domain I (amino acid residues 171-328) contains 4 α-helices and 5 β-sheets. Domain I has two ATP- binding motifs, Walker A and Walker B. Domain II (amino acid residues 329- 482) are composed of 3 α-helices and 8 β- strands. An arginine finger in motif VI in domain II is thought to be crucial for Nucleoside triphosphate (NTP) hydrolysis. Domain III (amino acid residues 483-619) comprises of 7 α-helices and 2 β-strands.6
Presently, novel drug inhibitors of JEV have been worked upon in recent times. Flavonoids namely (Kaempferol, Daidzein) and plant secondary metabolite like 4-hydroxyl panduratin A has proved to be effective inhibitors against JEV.11,12 Literature review has shown that Flavonoids and Isoflavones (class of phytoestrogens) inhibit virus infectivity. Flavonoids affect virus binding to cell membranes, entry into the cell, replication, viral protein translation within the host cell and formation of certain glycoprotein complexes of the virus envelope.13 The induction of certain transcription factors and secretion of cytokines; most of these effects have been attributed to a reduction in protein tyrosine kinase activity in treatment with genistein.14 In silico screening of 4-hydroxypanduratin A showed anti-viral activity against JEV NS2B/NS3 protease.12 Besides these compounds, antibiotic group compounds (Tetracycline and Aminoglycosides) have shown promising effects against flaviviral diseases. Broad-spectrum antibiotics like Minocycline tetracycline inhibited JEV and West Nile virus (WNV) induced infection in experimental models.15,16 Antiviral efficacies of two antibiotics (tetracycline and rolitetracycline) were found against Dengue (DEN) type 2 in a computer simulation that subsequently coordinated with in vitro experiments.17 Since the drugs have proved effective against other flaviviral etiologies (DEN and WNV), we hypothesized the drugs can be an ideal therapy candidate for JE too. This concept made the use of structure-based virtual screening (VS)/in silico to identify inhibitors of JEV plausible.
Therefore, in the present study, virtual screening was performed using iGEMDOCKv.2.1 (Generic Evolutionary Method for Molecular Docking) (software reference) to study the possible candidate compounds against JEV. This software is a graphics-automatic drug discovery system that involves post-screening analysis with pharmacological interactions, which could be used for integrating docking with the candidate compounds. The drugs were retrieved from NCBI Pubchem Compound database for docking at specific sites within NS3 helicase/NTPase domain of JEV protein. The selected drugs were then screened; post-analysis, and visualization of JEV NS3 helicase domain with compounds were performed. Based on docked conformations of tested compounds, we have proposed a model for inhibition of JEV NS3 protein. The NS3 protein may serve as a possible therapeutic drug target. Our findings may help to suggest candidate compounds as a possible therapeutic remedy against JEV.
The virtual screening method comprises four phases based on high-throughput molecular docking methods and crystal structures of target proteins. These phases include-target protein preparation, ligand preparation, molecular docking of the receptor protein with the ligands and post-docking and screening analysis.18
Preparation of target protein and ligand selection
JEV NS3 helicase/NTPase structure (PDB ID:2Z83) was retrieved from RCSB Protein Data Bank (PDB). The molecule was prepared for docking protocol initiation. All bound and unbound water molecules, ligands, and other organic compounds were removed. Hydrogen atoms were added and energy minimization on the entire three-dimensional crystal structure was performed. Energy minimization, receptor sites identification, and determination of Ramachandran plot to analyze the stability of the structure were performed using Swiss PDB Viewer 4.10. For ligand preparation, 10 candidate compounds were selected for the study. The group of compounds selected belonged to Aminoglycoside and Tetracycline group of compounds. Selection of the compounds was done by referring to the literature. Two standards, Ribavarin, a nucleoside antimetabolite and Minocycline tetracycline, a broad spectrum antibiotic was taken as standard drugs in our all experiments. All candidate compounds were retrieved from NCBI PubChem compound database. The compounds were obtained in MDL MOL format (.sdf) which depicted 3D conformations. Hydrogen atoms and Gasteiger charges were added into the ligands by using UCSF Chimera 1.8 stand-alone software (reference).Virtual screening
The iGEMDOCKv2.1 software provides a graphical environment for recognizing pharmacological interactions and virtual screening. Virtual screening of entire NS3 helicase domain with a library of 10 compounds was performed. The integrated docking, screening, post-analysis and visualization were performed [using RasMol (RasWin Molecular Graphics) software].Docking/docking analysis
iGEMDOCK v2.1 was used to perform molecular dockings. The core of iGEMDOCK v2.1 is the GEMDOCK [19]. The program utilizes a Generic Evolutionary Method for molecular docking and an empirical scoring function. Using iGEMDOCK v2.1, the predicted poses generated were directly visualized by a molecular visualization tool and analyzed by post-analysis tools.Post-processing of docking results and scoring function
iGEMDOCK v2.1 provides the post-analysis tools by using k-means and hierarchical clustering methods based on docked interactions and atomic compositions. The pharmacological scoring function of the docked pose is given as: Epharma= EGEMDOCK + E (E)pharma + 2 E (H)pharma +0.5 E (V)pharmawhere EGEMDOCK is the docked energy of GEMDOCK and E (E)pharma, E (H)pharma and E (V)pharma are the pharmacological scores of electrostatics, hydrogen- bonding and Vander Waal’s interactions respectively.19
The rationale behind this study
Based on previous literature, contrary to popular beliefs, antibiotics were also effective against arboviral infections. The mode of interaction of candidate ligands and JEV receptor protein has not yet been evaluated. Therefore, in present research work, we tried to perform virtual screening of selected antibiotics compounds all of which were coincidentally protein synthesis inhibitors against bacterial organisms. We selected JEV NS3 helicase/NTPase NS protein to perform docking with the selected candidate drug compounds. iGEMDOCK, a generic software was utilized to perform docking studies and computational analysis of the docked interactions.
Virtual screening for inhibitors of NS3 protein/Binding sites analysis
We evaluated antibiotic candidate compounds interactions with JEV NS3 helicase/NTPase structure (Figure 1). Chain A of JEV NS3 protein encodes a total of 448 amino acid residues.6 We performed virtual screening on NS3 protein with a screening set from NCBI PubChem database that contained 31 million compounds. The candidate compounds belonged to Antibiotics medications of Aminoglycosides and Tetracyclines groups. The selected candidate compounds were between 200 and 500 Daltons. In the present study, we had taken two standards for results comparison. The first one was Ribavarin, a nucleoside analog compound that is used against flaviviral infections.20 Hence, the drug was considered as a standard in our experiments. Minocycline tetracycline, a broad spectrum antibiotic compound belonging to polycyclic naphthacene carboxamide category was taken as the other standard. Minocycline was proved effective against JEV in experimental mice model and also against another flavivirus WNV in vitro.21,22 We used scoring values of both the empirical scoring function and pharmacophore-based scoring function as ranking conditions to score the reactive candidate compounds active with JEV NS3 protein. The compounds were then clustered by a hierarchical clustering dendrogram. The structures were grouped based on structure similarities, docked positions and protein-ligand interactions (Figure 2).
Computational analysis of antibiotics- NS3 protein interactions/ Post-docking analysis and hierarchical clustering of compounds
Docking of configured Antibiotic compounds with NS3 helicase/NTPase was performed. The docked conformations of the antibiotics derived inhibitors. Docking analysis showed that Kanamycin (carbohydrate based antibiotic compound that belongs to Aminoglycoside category) and tetracycline (derivatives of polycyclic naphthacene carboxamide category) group compounds showed the minimum binding energy that indicated strong binding affinity between the ligand moieties and viral protein. The results of docking runs are given in Table 1. The compounds were arranged in increasing energy or having a low binding affinity. It was observed that Kanamycin showed the highest molecular interaction energy. The hierarchical modeling illustrating the clustering of ligand and protein interactions are given in Figure 3. The docking conformations of the ligands are visualized in Figure 4. Majority of bond interactions were intermolecular van der Waals bond {strength (KJ/mol)-0.4-4.0 and distance (nm)-0.3-0.6} and a few hydrogen bonds {strength (KJ/mol)-12-30 and distance (nm)-0.3}.23 Majority of van der Waals contacts are formed between carbon, oxygen and nitrogen atoms. These three atoms resulted in 15 potential combinations which covered all van der Waals contacts between protein and compounds.
Compound |
Energy (Total) |
VDW (Van der waals) |
H Bond (Hydrogen bonding) |
Elec (Electrostatic energy) |
2Z83(mod)Kanamycin |
-147.367 |
-147.367 |
0 |
0 |
2Z83(mod)Rolitetracycline |
-124.552 |
-84.2985 |
-40.2532 |
0 |
2Z83(mod)- Doxycycline |
-121.816 |
-121.816 |
0 |
0 |
2Z83(mod)-Demeclocycline |
-118.599 |
-92.3541 |
-26.245 |
0 |
2Z83(mod)-Chlortetracycline |
-116.973 |
-116.973 |
0 |
0 |
2Z83(mod)- Minocycline |
-115.024 |
-94.8825 |
-20.1417 |
0 |
2Z83(mod)- Brefeldin A |
-86.9253 |
-64.8397 |
-22.0856 |
0 |
2Z83(mod)- Ribavarin |
-75.8248 |
-75.8248 |
0 |
0 |
2Z83(mod)- Gentamicin |
-58.9329 |
-58.9329 |
0 |
0 |
2Z83(mod)- Tetracycline |
117.823 |
130.609 |
-12.7856 |
0 |
Table 1 Energy of fitness of the different ligands with NS3 helicase/ NTPase in increasing order of energy
The major interacting residues present in NS3 helicase/NTPase which displayed conserved interaction were Lys200 (side chain), His268 (main and side chain), Asp291 (side chain), Gln457 (side chain), Glu491 (side chain), Asp542 (main chain and side-chain), Leu543 (main chain), Val545 (main chain), Arg600 (main chain and side chain), Val601 (main chain), Asp604 (side chain), Gln606 (main chain and side-chain), Ala607 (main chain and side chain) and Trp610 (side chain). All these residues were found to interact with the drugs through Vander Waal’s forces. Vander Waal’s force was the dominant part that contributed to 69% of binding energy. However, Phe326 (main chain), Asp328 (side chain), Ser329 (main chain and side chain), Ile333 (main chain) and Asp335 (side chain) interact with NS3 by hydrogen bond. Analysis of hierarchical dendrogram showed that bonds were formed in all the three domains- I (18), II (26) and III (13). Amino acid residues in motif II were shown to be essential for RNA helicase activity.24,25 The amino acid of Lys200 in walker A motif interacts with phosphates of ATP. The side chain of Lys200 plays an important role in electrostatic interaction with the substrate in NTP hydrolysis with an adequate charge and length to fit with the substrate. Also, Gln457 of motif VI present in ATP-binding pocket of JEV helicase participate in enzymatic activities of JEV helicase by interacting with the substrate. Amino acids in motifs I and VI participating in ATPase/helicase activity are important for viral replication as was reported in JEV and DEN.6,26 In our experiments, it was observed that the most three promising candidate compounds- Kanamycin (aminoglycoside), Rolitetracycline (tetracycline derived drug) and Doxycycline (Oxytetracycline derived drug) interacted with NS3 helicase with Lys200 and Gln457 amino acid residues among other interactions. The binding affinity of these three drugs with the two standards taken in our experiment- Ribavarin and Minocycline tetracycline was observed. Both the standards showed comparatively lower binding affinity. Therefore, based on our findings, we can state that the candidate compounds were better inhibitors of JEV NS3 helicase/NTPase protein compared to the standard drugs.
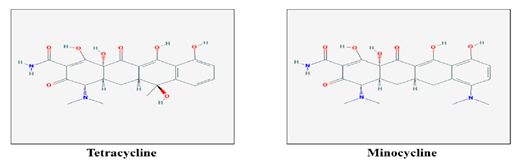
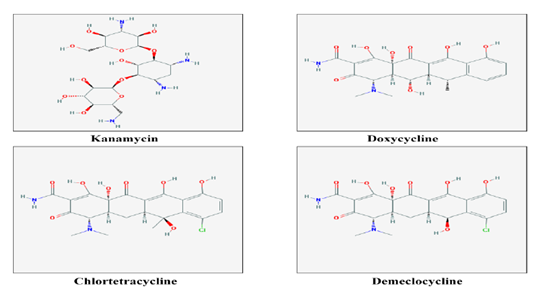
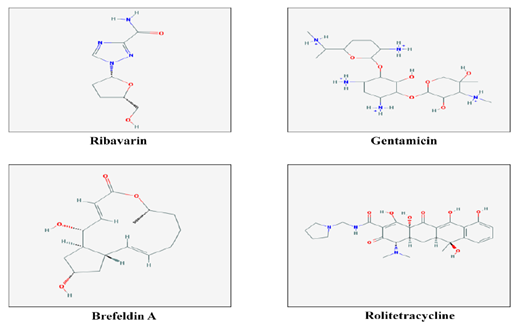
Figure 2 2-dimensional structures of 10 chemical compounds obtained from NCBI-PubChem Compound database to be used as ligands for screening and docking.
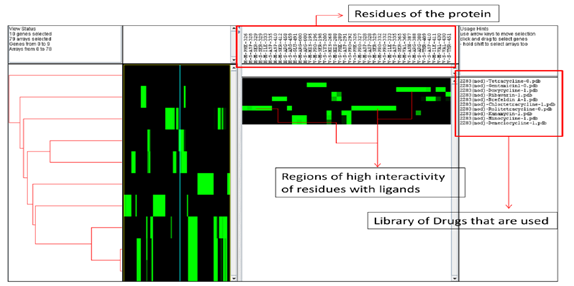
Figure 3 Java Tree View of the 10 ligands and the interacting residues of the protein obtained after virtual screening analysis using iGEMDOCKv2.1.
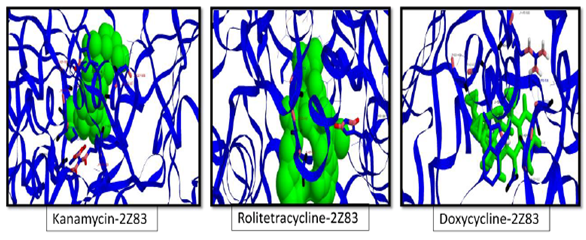
Figure 4 The three docking conformations (from left to right) of NS3 helicase/ NTPase (2Z83) with the 3 best ligands in terms of energy. The conformations are generated on the basis of total pharmacological energy calculated using the pharmacological scoring function of iGEMDOCKv2.1. (All the ligands are colored in green, carbon atoms in blue, oxygen atoms in black and nitrogen atoms in pink and hydrogen atoms in red).
At this junction, we can also state that possibly, the drugs have acted against viral replication of JEV as the probable mode of action. However, we can conclusively state that these in silico findings have paved the way of suggesting antibiotics as an alternative therapeutic option against JEV; however, the in silico results have to coincide with in vitro and in vivo experiments. These works remain to be performed ahead in the future.
In our research, we found that the candidate antibiotic compounds had shown bond interactions with JEV NS3 helicase/NTPase protein. Aminoglycoside- Kanamycin, and tetracyclines- Rolitetracycline and Doxycycline showed the most promising results. Interacting ligands docked with minimum energy that contributed to highly stable conformation. Bonds were formed in all three domains of the NS3 helicase. Maximum bonds were formed in domain II. Few prominent bonds were formed with amino acid residues in motifs I and VI that played a vital role in viral replication. These virtual screening showed that the candidate drug compounds were effective inhibitors against JEV by a probable process of acting against viral replication of JEV. However, the exact mode of action of antibiotics remains to be studied. We have to further study the efficiency of the candidate drugs in vitro and in vivo experimental models that will explore the horizons of drugs acting against JEV disease.
We extend our gratitude towards the Bioinformatics Centre (DBTBIF funded) of Gurucharan College, Silchar for providing necessary support for the overall undertaking of the project.
The author declares that there is no conflict of interest.

©2018 Nath, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.