MOJ
eISSN: 2374-6920


The p53 tumor suppressor protein, whose mutation has been implicated in some half of all human cancers, adopts a homotetrameric quaternary structure as the biologically active form. While much is known about the monomeric structure of the DNA binding domain, two competing models of the complete tetramer differing in the relative orientation of the DNA binding interfaces prevail in the literature. In this study we investigate the plausibility of an alternate quaternary structure assembled by overlaying four copies of the full length monomer as completed by molecular modeling to the four fragments constituting the crystallized tetramerization domains. On the basis of this modeling we discuss the plausibility of an alternate quaternary structure of p53 as well as explore implications for the biological role of p53 as a major transcriptional regulator of the cell.
Keywords: P53, tumor suppressor protein, P53 tetramer, cancer, protein folding; protein modeling, DNA binding, genome
The p53 protein, called the ‘guardian of the genome’,1 functions to integrate status information regarding the cellular integrity at key points throughout the cell cycle; when the genome suffers from stress such as oxidative damage, p53 activates repair pathways, and when the cell is deemed beyond remediation, it induces apoptosis to remove the damaged cell from the gene pool.2–5 Loss of native function of this protein has been attributed to some 50% of all human cancers.6 As a transcription factor, p53 effects these cell fates by discriminately binding to the promoters of the target genes required for the requisite status. As such, the interaction interface between p53 and DNA plays an integral role in determining the specificity of the interactions of the factor with its cognate binding sites7–9 and therefore in the proper determination of cell fate. Alterations of the p53 sequence causing changes to the DNA binding domain constitute a major mechanism by which mutations lead to cancer10–12 and mutations to this protein have been attributed to some 50% of all human cancers. Therefore, elucidating further detail of DNA recognition constitutes important groundwork required for the development of effective therapeutics designed to restore native function to mutant p53.
While a considerable amount is known about the structure of the monomeric DNA binding domain of p53 and its interaction with consensus sequence DNA at the atomic level of detail, the tetrameric form, generally accepted to be the biologically active form,13–15 has not yet unequivocally been determined.16 The DNA binding interface has been characterized from the pioneering x-ray crystallographic studies (1TUP),17 but the carboxy terminal site of tetramerization was not determined. However, the structure 1C2618 does capture the tetramerization of four short carboxy terminal fragments as a helix bundle and suggests a possible orientation for the tetramer. Another source of crystallographic information regarding the p53 protein comes from the interaction of a fragment with its regulator, MDM2. The purpose of this protein is to negatively regulate p53 action,19 which occurs by direct binding of the MDM2 to the N-terminal peptide of p53.20–22 The interaction has been captured crystallographically in the 1YCR structure in which the full length MDMD interacts with an alpha helical 13 amino acid N-terminal p53 fragment.23 In the literature, the two prevailing models of the full length tetramerized structure concur that the DNA binding interface of the monomer remains the site of interaction. Both models also presume that two p53 monomers first dimerize, and then a pair of dimers then dimerize. However, the proposed structures differ in the relative orientation of the binding interfaces due to different quaternary organization. Evidence from cryo electron microscopic data at 13.7Å resolution presents the structure as two oppositely faced dimers.24 The other model,13 put forth from crystallographic studies, suggests that the dimers bind along the DNA in a head to head fashion. These two models imply differences in how the four approximately 10 base pair contact sites of the monomers are arranged. The former model suggests that the DNA is contacted in two distinct locations, one for each dimer, separated by a possibly variable spacer such as a looped DNA. This model would also allow for two separate strands of DNA to be bound, one on each side. In contrast, the second model implies that both dimers bind with adjacent interfaces. While a consensus sequence of DNA interacting with the DNA binding domain has been empirically determined,8,25 our lab’s recent sequence analysis of genome-wide DNA binding sites26 indicates that many sites which in fact p53 do not follow the consensus, confounding the ability to distinguish between binding mechanisms by inspection of the sequence. Gaining a deeper understanding of the quaternary structure will thus aid in a deeper understanding of the protein-DNA recognition, suggest how that may factor into the effects of mutations, and inform the rescue of mutated p53 proteins by small molecule therapeutics.
The present study aims to investigate whether a quaternary structure based on the conformation suggested by the 1C26 structure of the carboxy termini would be plausible as judged by the resulting spatial arrangement of the tetramers and concurrence with the available crystallographic fragments. In the absence of a full length monomeric structure, the Phyre2 protein folding algorithm was implemented to generate a predicted structure. Four copies of the this modeled protein were structurally aligned to the tetramerized carboxy terminal crystal structure to yield a three dimensional quaternary representation of the p53 tetramer, which has been interpreted in terms of implications for its possible mechanism for DNA recognition and how an interchange between different tetrameric conformations could contribute to regulation of DNA binding.
Validation of full length p53 monomer sequence via sequence alignment
In order to obtain the tetrameric structure of human p53, the full length monomer was constructed as the first step, and then four copies were aligned to the tetrameric helix bundle to yield the structural model. The full length human p53 sequence (accession number P04637) was obtained from UniProt.27 To assess which portion of the full sequence aligned to the structurally determined 1TUP core domain,17 the tetramerization fragment of the helix bundle structure,18 and the N-terminal helix bound by MDM223 the respective FASTA sequences were downloaded from the PDB28 and a multiple sequence alignment was performed using Clustal Omega.29,30 Because the sequences have 100% identity, no sequence ambiguities arose, and the full length human sequence P0463 was expected to have a similar structure to the crystallized core and tetramerization domain. The UniProt FASTA sequence P04637 was thus used for the subsequent folding step.
Protein folding with Phyre2 and assessment of the folded monomer
The full length human p53 amino acid sequence was submitted to the Phyre 2 server (Protein HomologY/analogy Recognition Engine V 2.0).31 Both the normal and intensive modes were tested. The 1TUP chain B, the largest and most important p53 fragment crystallographically available, and the Phyre folded structures were locally aligned by with PyMol32 based on the selection of alpha carbons of all common residues, and the RMSD was used as a metric to gauge similarity; due to the identical sequence composition, the folded structure was expected to have high structural similarity. Because the normal modeling mode produced results which were incomplete, the intensive mode was implemented, and the highest scoring structure was chosen for further analysis. Molecular graphics were also generated with Pymol32 or the Visual Molecular Dynamics package.33
Assembly of the tetramer
The biological unit of the 1C26 crystal structure18 was obtained by replicating the crystallographic unit cell through four translations in PyMol to produce the four helix bundle. The full length intensely modeled Phyre2 predicted p53 monomer structure was quadruplicated, and the carboxyl termini of each was locally aligned by common alpha carbons to one of the four tetramerization domain fragments in the helix bundle of the 1C26 crystal structure analogous to the alpha carbon superpositions described above.
Sequence alignment
To elucidate the quaternary structure of the biologically active tetrameric form of p53, molecular modeling was undertaken to engineer a possible relevant structure based on the orientation suggested by the crystallized tetramerization domain fragments. Prior to structural alignment, extent of sequence overlap of the available relevant crystal structures with the full length human sequence was established by computing a multiple sequence alignment (Figure 1). The perfect sequence homology between the fragment sequences and the full length human p53 indicated that the available structures are relevant to the human sequence of interest. Furthermore, the alignment shows that 93 N-terminal residues including the proline-rich region require modeling, as do the 12 residues between the end of 1TUP and the start of the tetramerization structure, and 37 C-terminal residues; these do not have corresponding residues in the solved structures.
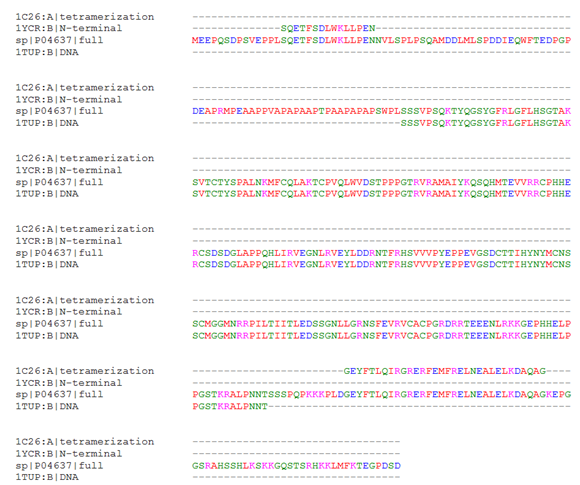
Figure 1 Sequence Alignment of Full Length p53 and Relevant Crystallized Fragment Sequences. The full length human p53 sequence from UniProt, P04637, was aligned to the tetramerization domain (1C26), the N-terminal helix interacting with MDM2 (1YCR), and the DNA binding domain (1TUP). Regions where P04637 aligns with another sequence indicates fragments for which structural information is available, whereas alignment with the gap symbol only indicates a lack of structural information.
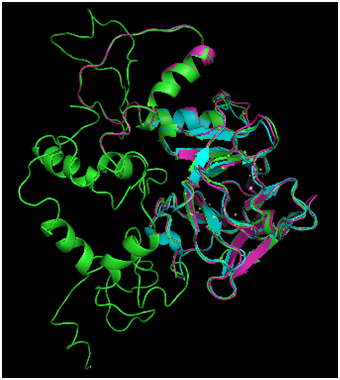
Figure 2 Comparison of Phyre2 Modeling Modes. Phyre structure prediction was carried out in normal (magenta) and intensive (green) modes. For comparison, the 1TUP DNA binding domain is also displayed (cyan). The normal mode produced an incomplete structure, whereas intensive produced a prediction for the full length p53 sequence.
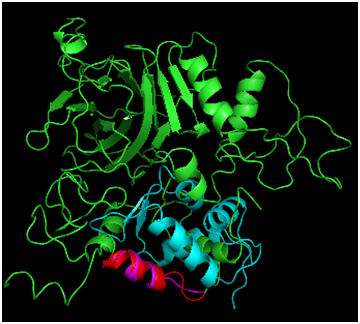
Figure 3 Alignment of Phyre2 Folded p53 to MDM2. Full p53 is shown in green, and MDM2 is shown in cyan. The p53 bound helix from the MDM2 crystal (magenta) was compared with the folded prediction (highlighted in red).
Optimizing the phyre2 calculation and alignment with 1TUP DNA binding domain
Phyre2 protein folding of the p53 monomer, carried out in both normal and intense modes and results were compared with the 1TUP DNA binding fragment (Figure 2). The normal modeling (magenta structure) predicted the DNA binding domain (cyan structure) with a fair degree of accuracy (RMSD=0.701Å). However, the algorithm failed to produce a prediction for the sequence in its entirety. The intensive mode (green structure) did provide a complete structure, and the degree of agreement to the DNA binding domain was greater (RMSD=0.440Å). The high agreement between the core domains is not surprising given that the Phyre algorithm31 includes the use of threading, a methodology that involves searching the PDB for structures with high sequence homology and using those as a template for the folded structure, which is then subjected to energy refinement. The numerous p53 structures present in the PDB provide ample examples of the expected fold, resulting in a prediction highly agreeing with the known structures.
Assessment with binding to MDM2 N-terminal helix
To assess how well the Phyre2 algorithm predicted the structure of the N-terminal portion of p53 captured in the 1YCR structure bound to MDM2, the structural alignment of the two helices was carried out (Figure 3), resulting in an RMSD of 1.656Å. While this agreement is rather poor for the N-terminal fragment of only 15 residues, the alpha helical structure was captured. Furthermore, MDM2 binds the helix in the crystal structure and may therefore induce a structural perturbation, which could be reflected in the RMSD value. In support of this, the superposition indicates that the structures cannot directly superimpose, as a helix of p53 satirically interferes with the MDM2 structure, suggesting that some conformational change must occur during the binding process; the helix may rotate away from the p53 core domain to alleviate the unfavorable steric interaction. Further studies taking into account the dynamic energetic of the conformational change may further elucidate the mechanism for this dynamic process. Figure 4 addresses the alignment of the Phyre2 folded full length structure (green) with a monomer of the tetramerization domain to assess the plausibility of the proposed modeling scheme. Not shown is the simultaneous alignment with 1TUP protein, but the DNA (sticks representation) and required zinc cofactor (grey sphere) which serves a structural role of appropriately orienting the DNA binding interface34–36 are displayed from that file so as to situate the relative location of the tetramerization domain, which has also been aligned (yellow). The alpha helical portion has been reasonably well captured as has the turn (RMSD=1.887Å), bearing in mind that the helical bundle comprises solely four copies of the tetramerization fragment may again indicate that the benefit of the rest of the protein may adjust the structure. Of note is the fact that although the protein folding algorithm does not explicitly account for cofactors, the structure is in excellent agreement in that region, likely due to the nature of the template fitting approach which could implicitly capture the structure in the absence of explicit inclusion in the calculation because the zinc bound conformation would effectively be learned from the geometries of crystals which are mostly bound to zinc.
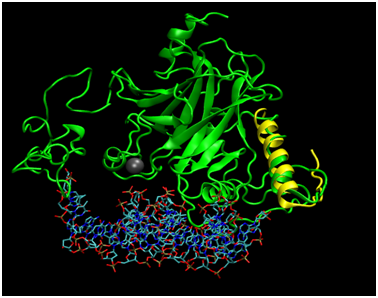
Figure 4 Alignment of Phyre2 Folded p53 with Tetramerization Domain Monomer. Full length p53 (green) has been aligned to the 1TUP structure (protein not shown) to display the relative position of Zn2+ cofactor as well the DNA binding site to the location of the tetramerization fragment as determined by an alignment to the 1C26 crystallized structure (yellow).
The structure of the modeled tetramer (Figure 5) resulting from this study indicates that construction of a tetramer model based on the tetramerization bundle yielded a plausible novel tertiary structure of the p53 tetramer in which each monomer occupies a separate volume. The orientation of the four monomeric chains suggests the dimerization of dimers, consistent with currently proposed models.13,24 The DNA recognition helices face towards a cleft between the subunits; the DNA could perhaps wrap around the structure, or bind in a loop formation. Perhaps the DNA is not simultaneously contacted by all four recognition helices simultaneously, which would help account for the lack of four strongly conserved DNA binding sequences in promoters. The separation of the DNA binding interface better coincides with the model purported by the cryo electron microscopy results. While a molecular comparison is not possible, the supposed structures do bear some degree of similarity, and this model suggests molecular coordinates that may account for the density viewed in the low resolution study. While molecular coordinates are provided with this method, bearing in mind that the structure has not undergone energy refinement to remove such energetically unfavorable interactions as van der Waal clashes has not been performed; such a study would benefit from a less crude alignment, perhaps with energy refinement as the structural alignment takes place. This could be carried out as part of a steered molecular dynamics simulation,37–39 and this coarse grain study suggests that obtaining a higher resolution tetrameric structure consistent with the conservation of the tetramerization domain as observed in crystallography is plausible.

Figure 5 Tetrameric p53 Generated from Superposition of Modeled Full Length Monomers to the Four Helix Bundle. Phyre2 folded full length p53 was replicated four times and each monomer was superimposed to one of the four components of the four helix bundle (1C26) crystal structure. Monomers are colored green, cyan, magenta, and yellow, and the four helix bundle is deep violet.
Further studies into the structure of the quaternary structure of p53 will elucidate how mutations on the surface of the monomer may affect the structure through interactions with other monomers; understanding the relative orientation of the interfaces with respect to one another can inform this type of study. Identification of the location of common mutations from the relevant literature that could disrupt the tetramer formation and map the mutations and/or mutation frequencies to the structure to study how these mutations may disrupt tetramerization. The mapping of these mutations on the constructed p53 tetramer would provide the ability to understand how mutations to p53 are situated with respect to the tetramerization interfaces. Additionally, these mapped mutations could reveal important insights about certain positions on the tetramer and could be influential in directing targeted cancer drug therapies.
Furthermore, bearing in mind that p53 is indeed a dynamic protein,36,40 an additional consideration regarding the tetrameric structure of p53 is that it may in fact interchange between more than one conformation. The Boltzmann ensemble of thermally accessible conformations could play a regulatory role, or help the protein switch between binding modes for discriminating between promoters. The studies to date have dealt largely with the unmodified side chains of the protein, but in fact, post translational modifications to p53 are known to play a role in switching p53 binding modes as part of the response to cellular status.4 That they mostly occur distal from the DNA binding region has been a puzzling observation, and little research by way of understanding the structural mechanism by which the modifications effect the binding specificity changes have been carried out. One possibility is that modifications could cause a structural reorganization in the quaternary structure by perturbing the relative stability of various substrates, which this model and those reported in the literature could represent. Further studies involving the characterization of the mechanism of how the post-translational code affects the molecular structure constitutes an interesting expansion of the p53 structural field for which the understanding of the tetrameric organization and possible conformational interchange may figure. As the quest for rational design of drugs to combat the formation of tumors progresses, gaining a deeper understanding of the dynamic nature of the protein and how it operates in a larger context will move the field towards the development of mutation specific drugs with fewer side effects and less undesirable cross reactions. The modeled tetrameric structure reported herein suggests a possible quaternary orientation of the p53 monomers in the tetramer consistent with the reported tetramerization domain and with similarities with the two major models currently debated in the literature. We have suggested that more than one model may simultaneously be correct, as the protein is highly dynamic. The structure informs researchers as to the relative location of the interfaces between the monomers as well as the relative orientation of the DNA binding domains, which can be used to better understand how mutations and therapeutics may affect the overall structure and therefore function of this very important protein responsible for safeguarding against the rapid production of damaged cells in the formation of cancerous tumors.
The authors graciously acknowledge the Vassar College chemistry department for the use of computational resources. KMT thanks Shelly Smith and Stephen M. Beare for technical support.
The author declares no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.