MOJ
eISSN: 2374-6920


With the advances in high-throughput genome/transcriptome sequencing technologies and mass spectrometry (MS)-based proteomics, thousands of gene-protein pairs can be matched and merged in a single experiment. It is of interest to perform a correlative analysis of gene and protein expression data and investigate the nature of their similarity/dissimilarity as it could harbour potential biomarkers or drug targets. Manual determination of data points of interest quickly becomes a very complex and laborious process. Thus, there is a high demand for automated ‘omics’ data integration tools that can not only routinely match and combine gene and protein expression values but also provide a measure to highlight meaningful biological insights. In this work, we applied a fast and easy approach to integrate large proteomic and transcriptomic data derived from the deep analysis of glioma cancer stem cells (GSCs). The proposed algorithm provides a mathematical distance between two data sets and asignes a direction of their interrelation based on the abundancies. We distinguished three types of the data correlation: concordant, anticoncordant where protein abundance was higher than that of the corresponding RNA and anticoncordant where protein abundance was lower. We investigated the nature of the observed discordances and were able to separate different, phenotypically divergent, classes of GSC lines.
Keywords: omics integration, proteomics, transcriptomics, bioinformatics, data similarity, genome, RNA, quantitative, non-correlation, stem cell, Uniprot, histocompatibility, Ubiquitination, proteoform
Proteomics, transcriptomics and genomics have a synergistic relationship, providing complementary information on genes and proteins associated with cancer or other diseases, metabolic and developmental states, and responses to drug treatments. Although proteomic analysis is dependent on genome completeness, it can also be viewed as a functional annotation tool,1,2 to uncover underlying biological mechanisms, discover novel genes3,4 and validate the presence of alternative spliceoforms.5 A combined approach enables comprehensive, multidimensional profiling of a complex biological system. However, establishing a correlation between proteomic and genomic/transcriptomic data is not a trivial process, in part due to the lack of appropriate bioinformatic tools. In a classic understanding of transcription regulation, one would expect a one-to-one ratio of gene and protein expression or for proteins to reflect the general RNA expression tendency: if an upregulation of a gene is observed, similar behavior is expected at the protein level. But even then, protein abundance represents combined abundances of all proteoforms in the sample. Additionally, proteins continuously undergo post-transcriptional6 and post-translational regulation, turnover, and directed ubiquitin-dependent degradation.7 Therefore, gene expression is rarely an accurate determinant of protein abundance.8 Even though higher levels of correlation between gene and protein expression have been reported,9 genes encoding histones10 and ribosomal proteins10,11 have been shown to correlate poorly with their respective proteins, due to the limitations of RNA-seq methodology. Additionally, when multiple Proteoform exist, each transcript would be assigned to the same gene, while proteomic tools would allow detection of each unique protein isoform, causing substantial discordances between data. It is of interest to highlight these classes of contradictory behavior in order to detect novel pathways of transcription regulation, disruption in transcription or directed degradation.
A straightforward approach is to create a scatter plot of transcript and protein quantitative values (i.e., abundances or p-value for significant changes) and manually assign the data points of interest. However, such an approach is very subjective and can be time-consuming in large high-throughput studies. Ideally, one would separate gene-protein pairs or subgroups in a robust, unbiased manner, and to perform statistical analysis of the results. For instance, k-means clustering as a first step of data handling was demonstrated to be a powerful classification tool when applied in temporal-based studies12 Through ‘omics’ integration we can differentiate data subgroups with various interrelationships: correlation, non-correlation or anti-correlation13 and treat them accordingly. Correlation coefficient, such as Pearson or Spearman, provides a value for each gene-protein pair that reflects one of these three interrelationships. However, this way the original relationship between protein and gene abundancies, which could harbor a key to understanding of their discordances, is lost. Alternatively, the number of dimensions could be collapsed to one by integrating the values. Although studies have been performed with the goal of creating a universal measure which would combine results from the different sources,14 the merging of opposing expression levels would average out final values. This could result in the elimination of key aspects in understanding of the regulation of cellular networks, for which both mRNA and protein expression is required.15
In order to compare and integrate values from multiple levels of expression, we have to acknowledge a number of limitations. Firstly, samples should be prepared identically to avoid the introduction of systematic biases at the later stages and to provide certainty that, when observed, differences and/or discordances are not artifacts of the sample preparation process. Ideally, a sample is divided into aliquots. This requirement is often not easily met due to limited sample amount, or when multiple laboratories collaborate. It is much easier to unify the process at the level of data processing, but the assumption needs to be made that samples were treated in the same way. Secondly, careful normalization and standardization has to be performed in order to bring protein and RNA expression values to the same level.
In this work, we explored a simple unbiased method to measure transcript-protein expression similarity. Using positive and negative signs for transcript-protein interrelation, we categorized gene-protein pairs and studied their behavior across multiple samples. We believe that, when meaningful, discordances occur systematicaly between cell lines. Here, we attempted to explain the nature of observed discordances and determine their effect on the data. We suggest that when transcriptomic and proteomic data are contradictory, we should consider dividing the data set into subgroups and process them accordingly to yield biological insights.
Proteomic data acquisition and pretreatment
Thirty-three cancer stem cell lines, provided by M.D. Anderson Cancer Center were divided in two aliquots for proteomic and transcriptomic studies. All proteomic samples were analyzed by use of nanoLC-MS/MS (Orbitrap Elite, Thermo) in a label-free quantitative proteomic workflow as previously described.16,17 Each block contained randomized groups of three GSC lines plus an external standard. Each sample was run in triplicate. The resulting .raw files were aligned by group in Progenesis LC-MS (Nonlinear Dynamics) and searched against a UniProt Human database (release September 2013) appended with a contaminant database (common Repository of Adventitious Proteins-cRAP http://www.thegpm.org/crap/) using PEAKS 6 (BSI). Due to the nature of further comparison raw intensities were used as quantitative measures. Peptide intensities were rolled up to proteins and processed further using DanteR. Protein intensities were log2-transformed and standardized with centering at zero.
Transcriptomics data acquisition and pretreatment
Paired-end whole transcriptome sequencing of 33 GSC lines, matched to cell lines for proteomics, was performed on the Illumina HiSeq platform after random priming and rRNA reduction. Each GSC line generated about 50 million paired-ends; each end was 75bp in size. Short transcript reads were mapped to 21,165 human protein coding genes in Ensembl reference transcriptome (ENSEMBL version 64). Downstream data analyses and RPKM (reads per kilobase per million reads) values were generated using Burroughs-Wheeler alignment, Samtools, and Genome Analysis Toolkit. More details on transcriptomic data acquisition are available in Lichti et al.16 Obtained RPKM values were then also log2-transformed and standardized with centering at zero. The resulting values were compared and integrated with the protein quantitative values and analyzed further using R.
Omics data integration and analysis
Each identified protein was matched with its corresponding gene name in transcriptomic data using UniProt.ws tool from Biocondutor, R (http://bioconductor.org/biocLite.R). Protein accession numbers were used as search keys to look up gene names. Within each cell line, genes/proteins with the same identifier were merged in one table, including genes/proteins that were not found in the corresponding dataset. Linear Euclidean distance is a standard dissimilarity metric used in hierarchical clustering. We utilized the same principal to estimate similarity measure between two datasets. In order to do so, corresponding protein and transcript expression values are assumed to be two points in the plane:

Where xi are protein quantitative values, yi-transcript’s values and n is a number of replicates. The resulting value is always positive, with a tendency toward smaller values when the original data is normally distributed. In the case when no corresponding gene or protein was found the distance value was assigned as N/A (not available). However, to reflect the directional relationship between datasets, +/- signs were associated with each value. All distances were assigned a positive sign when mean protein abundances were higher than those for corresponding transcript and negative, where mean protein abundancies were lower than the mean transcript levels. This created a normal symmetrical distribution around zero (Supplementary Figure 1). After removing genes that were not present across all cell lines, resulting values were used to cluster all cell lines and genes/proteins in a heatmap (Euclidean distance metric, Ward linkage metric, Mass Profiler Professional, Agilent, version 12.6.1) and to track the enrichment for a specific characteristic (function, localization or sequence motif).
Functional annotation
Subgroups of gene-protein pairs with anticoncordant behavior were annotated based on their Gene Ontology (GO). QuickGO (http://www.ebi.ac.uk/QuickGO/) was used to extract biological functions and cellular components for each gene/protein of the interest. Genes/proteins involved with the same functional annotations were summed in each cell line separately and then combined across all the cell lines in one master table. GO terms were then clustered based on their protein count using k-means (with 5 centers) to determine the major processes where discordances occur, cluster with the highest protein counts across all cell lines for each GO term. Distances for transcript-protein pairs from each GO group were extracted and compared with the overall distribution in a histogram. Histograms were generated using R to overlay the distribution of distances for all proteins vs. the ones annotated with a specific GO term (Supplementary Figure 2). For each matched protein, its sequence in fasta format was downloaded from UniProt website (http://www.uniprot.org/) and mined for degradation motifs. Simple regular expression description was used to recognize “destruction box” (R-x-x-L-x-x-x-x-N/D/E, x-any amino acid),18 “KEN box” (K-E-N-x-x-x-N/D),19 PEST region (P-E-S-T)20 and N-terminal stabilizing and destabilizing amino acids (M/S/A/T/V/G and F/L/D/K/R respectively).21 To identify ubiquitylated proteins we matched our dataset to the entire list of experimentally identified ubiquitylated proteins from UbiProt22 (137 proteins in human). All the analyses and plotting was performed in R.
Previous works in the area of ‘omics’ integration proposed algorithms that treat all transcriptomic-proteomic data in the same way.14 We suggest that not all of the expression values should be processed in the same manner. In order to yield biological insight, meaningful information must be separated from technical bias. Here, for each RNA-protein pair, we utilized a simple and robust data similarity metric that is commonly used as a first step in hierarchical clustering to distinguish concordantly expressed pairs of transcripts and proteins from discordantly expressed ones. Naturally, the resulting distance measure is always a positive value. However, to reflect the interrelationship between RNA and protein expressions, we assigned +/- signs to all the distances. As a result, the values were normally, symetrically distributed around zero (Supplementary Figure 1). This allowed us to visually separate three gene-protein groups for further analyses: concordant where gene and protein expressions are equal, anticoncordant where gene expression is higher than protein (similarity measure below -2) and anticoncordant where gene expression is lower than protein (similarity measure above 2). We based a threshold assignment (-2, 2) on the distribution of distances in a sample.
Different ‘omics’ techniques operate with different databases and therefore different identifiers. In order to enable merging of the data, the identifiers must be unified. Transcriptomic analysis provides gene names, while proteomics yields protein IDs and accession numbers. Gene names are not unique; there is redundancy and even identical names between different species. Therefore, using gene names to look up protein IDs is suboptimal. On the other hand, protein accession numbers are absolutely unique and can be searched easily in UniProt to obtain corresponding gene names. The existence of numerous proteoforms as products of the same gene leads to a large number of redundant matches. Even though all of these proteins had missing values in >1 cell line, often, proteomic data derived from various proteoforms were complementary to each other. For instance, if one proteoform was quantified in only a subset of GSCs, another proteoform was usually measured in the remaining cell lines. However, it is not an ultimate rule and in the case of histocompatibility complex (HLA-A and HLA-B) the same proteoform would have various expression values in different cell lines. In those cases, each protein’s abundance is compared to the same gene.
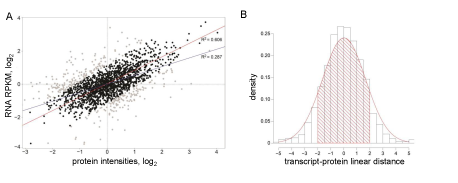
Theoretically, we expect RNA and protein abundances to be normally distributed along the line y=x which is reflected in the distribution of linear distances. Therefore, we used this line as a dissimilarity cut off. Experimentally, our analysis showed that 15% - 25% of proteins had discordant RNA expression in any given cell line, which resulted in poor protein-transcript correlation. Figure 1A illustrates a typical correlation plot generated from the data from one GSC line (see Supplementary Figure 3 for all GSC lines). These two datasets demonstrate low correlation (R2 = 0.287). When RNA-protein pairs with distance <-2 and >2 (Figure 1B) are removed from the set, R2 improved to 0.606. The choice of the threshold can be customized and, for the best result, should be sample specific.
Logically, all discordances occur due to two reasons: technical or biological. We hypothesized that a fraction of discordances could harbor keys to the understanding of disruptions in the regulatory pathways, so we focused our further analyses on the detection of such RNA-protein pairs. In those cases, we assumed that meaningful discordances should occur systematically at least in a subset of the cell lines. In order to test this hypothesis, we generated a heatmap of all non-N/A-containing transcript-protein distances across all GSC lines (Figure 2A). All distances with “+” sign were colored red, while all the “-“ distances were colored blue. Three clear clusters of genes could be noted in the heatmap: mostly negative distances across all cell lines, mostly positive and differential between cell lines. To visualize the effect of phenotypical characteristics on this clustering, we overlaid the Cancer Genome Atlas (TCGA) classification23 of the cell lines onto the heatmap. This classification distinguishes GSC subtypes based on the gene expression profile and original tumor clinical characteristics. Clusters enriched for either negative or positive distances only contain large subgroups of discordant RNA-protein pairs, values <-2 and >2 respectively (Supplementary Figure 2). In the differential cluster, a group of cell lines that belong to the classical subtype, except for two mesenchymal GSCs, appeared to have mostly negative distance values, while the rest have mostly positive values (Figure 2B). However, almost all genes/proteins that organize this cluster are concordant, values between -2 and 2 (Figure 2C). Cell line that appears as an outlier on Figure 2B (gsc275) is marked as proneural subtype; however, the tumor that this cell line is originating from is classified as classical. Majority of the classical cell lines that cluster together are also classified as such in the original tumors. A literature search on top 13 drastically different proteins Table 1 showed that majority of them are known to be associated with glioblastoma.24-35 Clinically, classical cell subtype is characterized as the most resistant to aggressive treatment.23 In the list of genes differentiating classical cell lines from the rest MARCKS30 and STMN125 are known to affect tumor survival and its resistance to drugs. Additionally, even though PSMA1 itself is not directly associated with glioblastoma, the other subunit of 26S proteasome PSMB4 was previously reported as one of the survival genes.36 Genes HDGF, MARCKS and STIP1 promote tumorgenesis26 and cancer cells proliferation.30,33
Even though only concordant values were characteristic for GSC phenotypic separation, it is possible that systematically discordant genes and proteins are not directly related to glioblastoma, but may be important in cancer development and otherwise be overlooked. In that case, we would expect discordant gene-protein pairs to fall into the same functional category or gene ontology (Supplementary Figure 4). To study the effect of function on expression correlation, we examined the distribution of gene-protein distances for top two specific GO terms, which were determined as the most represented across the majority of the cell lines after k-means clustering of all GO molecular functions for anticoncordant genes/proteins (see Methods). Ideally, if a function is enriched we would observe a shift in the distribution of distances. However, histograms for translation or regulation of transcription, the top molecular functions, did not demonstrate any discrete functional grouping.
Transcripts and proteins that are systematically discordant across multiple cell lines could either be a definition of a normal state of the cell or an artifact of data acquisition. The proteome represents a snapshot of the system, which partially consists of proteins that do not require frequent renewal, such as structural or membrane proteins. Because their synthesis happens at the early stage of cell life, these proteins might not be accurately represented by RNA-seq data. We tested that hypothesis for the subcellular localization (Supplementary Figure 4), and no specificity toward lower (in the case when these proteins are lost due to imperfect sample preparation in the proteomics experiment) or higher (when RNA-seq data is inaccurate) distances was detected. In fact, distribution of distances for a subset of genes/proteins almost perfectly mimicked the overall distribution for all genes/proteins, with the maximum frequency around zero and gradually decreasing towards the positive and negative ends.
We hypothesized that proteins with ubiquitin-mediated degradation or instability should demonstrate anticoncordant gene-protein relation where protein abundancies are significantly lower than those of corresponding transcript. Consequently, distribution of RNA-protein distances for those pairs would shift towards more negative values. Ubiquitination is a post-translational modification that directs substrates towards degradation and has been shown to frequently occur in cancer-associated proteins.37 It is impossible to detect ubiquitination at the RNA level. However, it is possible to predict its sites in the protein sequences. By searching UbiProt,22 a database of experimentally detected ubiquitylated proteins, we found 76 of those proteins in our dataset. When we examined the calculated distances of these proteins we observed no specific trend towards positive or negative values. In fact, there were barely any gene-protein pairs with distances below -2 (Supplementary Figure 5). We also searched for the other known degradation signals such as “destruction box”, “KEN-box”, PEST region and destabilizing N-terminal residues expecting transcript-protein pairs with large negative values (when protein abundancy is significantly lower than that of the transcript) being enriched. However, transcript-protein distances were normally distributed for each of the degradation motives and no correlation was shown (Supplementary Figure 6). Similarly, little to no dependence was observed when analyzing sequences with stabilizing N-terminal residues21 (Supplementary Figure 6). These findings may be partly explained by the fact that consensus sequences rules were very generic, resulting in a lot of bias, or there is indeed little effect of destabilizing motifs on the protein’s half-life and turnover.38
An alternate explanation is that ubiquitin-directed degradation resulted in protein amounts below the instrument’s limit of detection, and these proteins appear as N/A. Additionally, we compared protein stability predicted by ExPASy ProtParam Tool39 (http://web.expasy.org/protparam/) with transcript-protein abundancy distances. ExPASy ProtParam Tool computes structural instability index based on the proteins sequence, with <40 being stable. We anticipated that negative subgroup of distances would be enriched with unstable proteins, therefore explaining why higher level of RNA is observed compared to the corresponding protein. However, no conclusive correlation between those parameters was observed (data not shown), suggesting that the cause of these discordances are technical rather than biological. Such technical factors affecting transcript-protein correlation have been discussed in greater details by Ning et al.40 It is fairly safe to acknowledge that discordant RNA-protein pairs hold less biological interest compared to the concordant ones and occur mostly due to the technical limitations. We demonstrated that they could be detected in a large ‘omics’ data in a robust manner and be discarded from the data sets to improve their correlation. However, they may still contain valuable information to identify signaling pathways associated with cancer or other diseases. In a simple profiling experiment such as this one, we are limited to operate only with raw abundances that do not always reflect gene/protein behavior in a cell. Applying specific conditions, such as irradiation, for pairwise comparison (control-treatment) can potentially decrease the number of discordant gene-protein pairs, as they most likely will exhibit similar direction of expression changes in response to a stimuli,40 to reveal true relationship between transcript and protein expression (similarly to12) and more meaningful insights.
Gene Name |
Uniprot AC |
Protein Name |
Publications Associated with Glioblastoma |
PTMS |
P20962 |
Parathymosin |
1[41] |
RPLP2 |
P05387 |
60S acidic ribosomal protein P2 |
Associated with cancer |
SERBP1 |
Q8NC51 |
Plasminogen activator inhibitor 1, RNA-binding protein |
1[24] |
STMN1 |
P16949 |
Stathmin |
1[25] |
HDGF |
P51858 |
Hepatoma-derived growth factor |
4[26-29] |
MARCKS |
P29966 |
Myristoylated alanine-rich C-kinase substrate |
2[30,31] |
COX5B |
P10606 |
Cytochrome c oxidase subunit 5B, mitochondrial |
- |
MARCKSL1 |
P49006 |
MARCKS-related protein |
- |
ZYX |
Q15942 |
Zyxin |
zyxin family member TRIP6 [42] |
PGRMC1 |
O00264 |
Membrane-associated progesterone receptor component 1 |
- |
PSMA1 |
P25786 |
Proteasome subunit alpha type-1 |
1[35] |
ATP5H |
O75947 |
ATP synthase subunit d, mitochondrial |
- |
STIP1 |
P31948 |
Stress-induced-phosphoprotein 1 |
3[32-34] |
Table 1 Top 13 the most different genes/proteins that distinguish GSC lines based on their TCGA classification in Figure 2B

In our transcriptomic and proteomic data, 20% of proteins on average were discordant with RNA-seq amounts. We utilized linear Euclidean Distance to assess similarity between two types of ‘omics’ data in an easy and robust manner. Even though we demonstrated that discordant transcripts and proteins do occur systematically, these transcript-protein pairs could not be described by a specific rule and did not fall into one or a few functional categories based on GO analysis. Moreover, they could not be characterized by a specific quality, and most are probably a result of erroneous identification, sample preparation artifacts or due to measurement errors at the lower end of instrument’s level of detection. Removing those genes/proteins from the data set improved correlation between transcriptomic and proteomic data by 50%. Adding directionality to the similarity measure enabled us to distinguish three correlation classes and highlighted phenotypical differences between the GSC lines based on their TCGA classification.
Authors would like to thank Alexander Shavkunov and Huiling Liu for their technical assistance and data acquisition. We also would like to thank Akos Vegvari for the valuable comments and scientific discussion. Financial support from the Cancer Prevention and Research Institute of Texas and University of Texas Medical Branch (CLN) are gratefully acknowledged. This work was also supported by Guion Pool Keating Endowment Award (CLN). EPS was supported by the National Institutes of Health/National Cancer Institute (SPORE Grant No. P50CA127001, R01CA190121).
EM developed the concept and performed the analysis. CFL performed proteomics data pre-treatment and helped with postprocessing. QW performed transcriptomic data pre-treatment. EPS provided experimental materials, data and valuable input on the biological interpretation. CLN provided guidance, valuable discussions of the concept and interpretation of the results. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The author declares no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.