MOJ
eISSN: 2374-6920


Expression of Lympho-Epithelial Kazal-Type-Inhibitor (LEKTI), a broad spectrum protease inhibitor, is dysregulated in head and neck squamous cell carcinomas (HNSCC) and HNSCC cell lines. Here, we investigated expression of LEKTI in primary tumor specimens of 81 patients with SCC of the oral tongue in correlation with pathologic findings and clinical outcomes. IHC analyses have shown that LEKTI expression is negative in 31, intermediate in 44, and strong in 6 patients. Correlative analyses between LEKTI expression and perineural Invasion (PNI) and lymphovascular invasion (LVI) demonstrated that the relative risk of PNI and LVI were 3.2 (95% CI, 1.2 to 8.9, p=0.007 by Chi Square Test) and 6.0 (95% CI 1.2 to 40.8, p=0.01 by Fisher’s Exact Test) respectively in patients with LEKTI-negative tumors compared to those with LEKTI-positive tumors. Kaplan-Meier estimates showed that patients with LEKTI-negative expression had a 20% increased risk of disease recurrence (HR 1.19 and 95% CI 0.61 to 2.33, p=0.23 by Log rank test) and an 80% increased risk of death from all causes (HR 1.78 and 95% CI 0.34 to 9.41, p=0.48 by Log rank test). Analysis of the covariates for disease recurrence and death in tongue cohort found no significant differences in age, gender, T-stage, grade, N-stage, and postoperative treatment between patients with LEKTI-negative and LEKTI-positive tumors. Further, we present evidence that transcriptional regulation is a very likely mechanism accounting for loss of LEKTI mRNA and protein from HNSCC. These data confirm our previous in vitro and orthotopic model of tongue cancer findings and shed new light on mechanisms of PNI and LVI in HNSCC.
Keywords: SPINK5, LEKTI, lymphovascular invasion, perineural invasion, HNSCC, fisher’s exact test
LEKTI, lympho-epithelial kazal-type-inhibitor; SPINK5, serine protease inhibitor kazal-type 5; HNSCC, head and neck squamous cell carcinoma; PNI, perinural invasion; LVI, lymphovascular invasion
Local tumor invasion including perinural invasion (PNI) and lymphovascular invasion (LVI) occur by the attachment of tumor cells to components of the extracellular matrix (ECM) and by degradation of ECM by proteinase enzymes elaborated into the tumor microenvironment.1-6 These processes are regulated by such proteolytic enzymes as serine proteases, cysteine proteases, and matrix metalloproteinases (MMPs) tightly balanced by their endogenous inhibitors in the tumor microenvironment.7-13 Thus, inhibition of such proteinases can disrupt critical steps of invasion and metastasis. Indeed, several proteinase inhibitors have shown importance in a range of cancer types by the loss of expression correlating with advanced tumor progression.14-19 Moreover, proteinase inhibitors have been demonstrated to hold tumor suppressor functions that oppose tumorigenesis, proliferation, angiogenesis, invasion, and metastasis.20-22
An inhibitor of multiple serine proteinases, lymphoepithelial kazal-type inhibitor (LEKTI), was identified and cloned in our laboratory on the basis of its constitutive expression in normal oral mucosa and loss of its expression in matched head and neck squamous cell carcinoma (HNSCC) specimens and multiple HNSCC lines.23 Recently it was confirmed in a sub-set of HNSCC.24 It was also shown by several investigators that LEKTI protein was encoded by SPINK5 gene and mutations in SPINK5 has been linked to the inherited disorder known as Netherton Syndrome.25-41 We produced recombinant full length LEKTI and several of its fragments using baculovirus expression system and established that recombinant human LEKTI inhibits a battery of serine proteinases in vitro including plasmin, trypsin, cathepsin G, human KLKs, and elastase, enzymes implicated in the activation of MMPs.42-47 Recently, we demonstrated that stable expression of LEKTI in HNSCC OSC-19 cells resulted in markedly decreased levels of expression of genes encoding MMP-9, MMP-14, KLK5, and ADAM8. Furthermore, LEKTI over expressing cells displayed striking morphological changes are more adhesive and less invasive,48 These results demonstrate a novel negative regulatory role for LEKTI in modulating the production of key MMPs involved in ECM degradation and suggest that loss of LEKTI in HNSCC tumor cells could have a pivotal role in HNSCC progression. Subsequently, we determined the consequences of LEKTI re-expression on the in vivo changes in the tumor growth and invasion using an orthotopic model of tongue cancer.49 We have demonstrated that in the tongue tumors of mice, lymphovascular invasion or perineural spread was found in 100% of tumors derived from vector or parental cell lines but was almost totally absent in all tumors derived from LEKTI-expressing clones. Thus, we hypothesized that loss of LEKTI expression in primary tumors correlates with aggressive biologic behavior in patients with HNSCC. To test our hypotheses, we now investigated the expression of LEKTI in primary tumor specimens of patients with SCC of the oral tongue in correlation with pathologic findings and clinical outcomes and also the mechanism of its loss. Altogether, our results showed a heterogeneous expression of LEKTI, an association between LEKTI expression and occurrence of PNI and LVI and an epigenetic mechanism of regulation in SCC of the oral tongue.
Materials
The following reagents were obtained commercially as indicated: Erase-a Base nested deletion mutagenesis system, luciferase reporter plasmid pGL3-Basic and Dual luciferase assay kit (Promega Corp., Madison, WI); pCRII-TOPO (Life Technologies, Rockville, MD); Genomic DNA isolation kit (Qiagen, Valencia, CA ); sodium bisulfite, hydroquinone, sodium acetate, 5'-azacytidine (Sigma-Aldrich, St. Louis, MO); primary normal human epidermal keratinocytes (NHEKs) and keratinocyte growth medium (Cambrex Biosciences, Walkersville, MD); precast sodium dodecyl sulfate (SDS)-polyacrylamide gels, prestained markers (Bio-Rad Laboratories, Hercules, CA); anti-LEKTI mAb 1C11G6 (Zymed Laboratories, San Francisco, CA); horseradish peroxidase-conjugated goat-anti-mouse IgG (H+L) (Jackson ImmunoResearch Laboratories, West Grove, PA); lipofectamine 2000 and pcDNA3.1 (-) (Invitrogen, Carlsbad, CA); ECL kit (Amersham Bioscience Corporation, Piscataway, NJ); Kodak X-AR5 films (Eastman Kodak, Rochester, NY); Sense and antisense oligonucleotides specific for human LEKTI were synthesized by Genosys Biotechnologies Inc. (The Woodlands, TX); restriction endonucleases and polymerase chain reaction reagents (New England Biolabs, Beverely, MA); UMSCC-1 is an established HNSCC tumor line derived from patients with squamous cell carcinoma of the floor of mouth.
Cell culture and transfections
The HNSCC cell line UMSCC-1 was obtained from Dr. Tom Carey at the University of Michigan. NHEKs were cultured in keratinocyte growth medium containing low-calcium. All cells were cultured at 37°C in humidified incubator with 5% CO2 and 95% air. Cells are co-transfected in triplicate with 1mg reporter construct and 25ng Renilla luciferase or control vector plasmid DNA using Lipofectamine 2000 according to the manufacturer’s instructions. Approximately 50% transfection efficiency was achieved as determined by transfection with a GFP control plasmid.
IHC of LEKTI protein expression in primary tumors
The primary tumor specimens were recut and stained with a purified mouse anti-LEKTI monoclonal antibody. Specimens were stained for LEKTI with an automated stainer. Briefly, sections (4µm thick) will be deparaffinized, dehydrated, antigen retrieved using microwave methods, immersed in 0.3% H2O2 for 10min at room temperature in methanol, and washed with PBS. The sections will be incubated with 3% BSA in PBS, washed twice, and then incubated with a mouse monoclonal anti-PCNA antibody (Dako) for 1 h at room temperature. Primary antibody will be detected with a Vectastain ABC-alkaline phosphatase kit (Vector Laboratories, Burlingame, CA). Two investigators will examine the stained specimens independently. In each case, three arbitrary separate microscope fields (×200) will be examined to count immunoreactive tumor cells and the total number of tumor cells. The average percentage of immunostained cells will be defined as the labeling index (LI) and used for statistical analysis. The1C11G6 mAb, which is specific for LEKTI, will be detected using the Vectastain ABC-alkaline phosphatase kit according to supplier's instructions. Slides were reviewed and categorized by two independent investigators. LEKTI scoring was done as follows: Tumors with weak to no LEKTI staining in >95% cells were considered negative; Tumors with moderate LEKTI staining in >5% of cells or strong staining restricted to more differentiated tumor were considered intermediate; Tumors with strong LEKTI staining in >5% of cells, not restricted to more differentiated areas were considered strongly positive. The surgical pathology reports were reviewed for histopathologic features of the primary tumor. Medical records were reviewed for covariate and clinical follow-up data. Time-to-event analysis was performed with the Kaplan-Meier method for patients stratified by LEKTI-staining pattern.
Luciferase reporter assays
A series of 5' deletion mutants derived from the 343bp LEKTI promoter is generated using the Erase-a Base nested deletion mutagenesis system and cloned into the luciferase reporter plasmid pGL3-Basic. To control for efficiency of transfection, the pRL-CMV vector containing Renilla luciferase driven by the CMV enhancer/promoter is co-transfected with constructs derived from pGL3-Basic. The products of firefly and Renilla luciferase can be distinguished from each other using the Dual-luciferase Reporter Assay. Cells (4 ×105) are seeded in 6-well plates the day before transfection. On the day of transfection, 3µg DNA from each construct and 0.125µg pRL-CMV (internal control) is transfected into each well with 3µl Lipofactamine 2000. After incubating at 37°C for 4 h, complete medium is added and cells are harvested 40 h after transfection by scraping off into lysis buffer (500µl) as described in the Dual luciferase assay kit. After storing at –80°C for overnight, cell extracts are cleared by centrifugation for 2min at 4°C and 20µl are used for luciferase assay. The luciferase activity is measured with the dual-luciferase reporter assay system by a micro titer plate luminometer (Dynex Revelation 4.06, Dynex Technologies, Chantilly, VA) according to the manufacturer’s protocol. Transfections are performed in triplicate wells.
Primer extension
An antisense oligonucleotide approximately 100bp downstream of the predicted start of site of transcription is synthesized, labeled with T4 polynucleotide kinase and [γ-32P] ATP, hybridized to 20mg total RNA from NOE cells, and extended by AMV reverse transcriptase (in the presence of actinomycin D). The extended DNA product is treated with RNase, ethanol precipitated and loaded onto a 9% acrylamide/7M urea sequencing gel. To pinpoint the start of transcription, a 500bp fragment of genomic DNA containing the primer extension binding site and upstream genomic sequences is cloned ahead of time into a plasmid vector, and cycle sequenced with the same antisense oligonucleotide used in the primer extension experiment. The primer extension product is loaded alongside the sequencing reactions for comparison.
Northern blot and real time PCR
Total RNA isolation, Northern blot and Real-time PCR were performed as described earlier.28 Total RNA was prepared using TriZol reagent (Invitrogen) according to the manufacturer’s instructions. For Northern blot, 20µg total RNA was applied to a 1% formaldehyde agarose gel. After transferring RNA to Hybond-Nþ membrane (Amersham), the membrane and the filter were hybridized with 32P-r-human LEKTI or 32P-GAPDH. For Real-time PCR, 2µg total RNA were reverse transcribed (RT) by Superscript II (Life Technologies) in a 25µl total reaction volume containing RT buffer, random hexamers, dNTP, and RNase inhibitor (Roche Applied Science, Indianapolis, IN). Real-time PCR was performed in a 25µl total reaction volume containing 1µ of 1:10 diluted cDNA obtained from RT reaction, 12.5µl of TaqMan Universal PCR Master Mix without AmpErase UNG, and 1.25µl of specific primers for LEKTI gene on ABI Prism 7900HT. As a control, 18S primers were used, and cDNA was diluted to 1:500. Serial dilutions of the standard templates were also used for parallel amplifications. Levels of LEKTI mRNA were normalized to those of 18S in each sample.
Nuclear extracts isolation and DNAse i footprinting assays
Cells are scraped in cold PBS, centrifuged, and suspended in buffer A (10mM HEPES, 10mM KCl, 0.1mM EDTA, 0.1mM EGTA, 1mM DTT and 0.5mM PMSF, pH 7.9). After 15min on ice, cells are centrifuged and nuclear pellets disrupted in cold buffer B (20mM HEPES, 0.4 M NaCl, 1mM EDTA, 1mM EGTA, 1mM DTT,1mM PMSF and 20% glycerol, PH 7.9) by homogenizing with 10 strokes and incubating further for 30min at 4°C. After an additional centrifugation, the supernatant containing nuclear extracts is harvested. DNase I footprinting is carried out using Promega’s core footprinting kit with modifications. Probes are PCR generated, subcloned into the pBlueTOPO vector (Invitrogen), excised with HindIII/BamHI, gel purified using a QIAEX II Gel Extraction Kit (Qiagen), and labeled with [32P] ATP using T4 polynucleotide kinase. Labeled probe is digested with either HindIII or BamHI to achieve selective labeling of either sense or anti-sense strands. Increasing amounts of nuclear extract is incubated with labeled probe (10,000cpm) at room temperature for 10min, followed by digestion with 6 units of DNase I at room temperature for 3min. Digested products is phenol/chloroform extracted, ethanol precipitated, and analyzed using 6% acrylamide sequencing gels containing 8 M urea. For comparison, DNA-sequence reactions of the genomic DNA using a proximal primer are run in parallel.
Microdissection, DNA extraction, and LOH
Consecutive 5-micron sections from formalin fixed paraffin embedded specimens are cut and placed onto positively charged slides. One slide is stained with H&E, and another with Methyl Green. Areas of tumor or normal identified on H&E slide are used to localize corresponding regions on Methyl-Green stained slide to be microdissected. A number 15 scalpel blade is dunked into PCR/PK buffer containing 0.5% Tween 20 and solution transferred to area that is to be scraped. Approximately 100 to 200 cells are scraped into puddle of PCR/PK buffer and transferred to a PCR tube containing 15µl PCR/PK buffer plus Proteinase K (0.4mg/ml). Digestion reactions are overplayed with mineral oil and incubated at 55°C with shaking overnight. Fresh Proteinase K is added and the overnight incubation repeated two more times. Proteinase K is heat inactivated by incubation for 15min at 95°C, and 5µl digested DNA is used per PCR reaction. For LOH analysis, DNA is amplified using Taq Polymerase Gold (Perkin Elmer) for 37 cycles in the presence of fluorescently labeled primer. The size of PCR products is then compared on an automated DNA sequencer. In some cases, DNA is extracted from leukocytes using DNAzol (Molecular Research Products) according to the manufacturer’s instructions. Primer sets for the two polynucleotide repeat regions within the LEKTI locus are: (1) 5’CACAAACATACCAATTTTAACAAACC-3’(sense) and 5’-
TGAACCACATCGCTATTAAGAGTTTT-3’(antisense); and (2) 5’-CAGCCTGGGCAACAGAGTAA-3’(sense) and 5’-CACCACTGAATAGATTTTAGGGAGG-3’(antisense).
Bisulfite treatment and LEKTI promoter methylation
The methylsation patterns in LEKTI promoter region was determined as described in the literature.50 Genomic DNA from NHEK and UMSCC1 cells (2µg) was isolated and treated with bisulfite according to the published protocol.50 Bisulfite-treated genomic DNA (0.2µg) was used in PCR using bisulfite PCR primers designed by MethPrimer software from University of California San Francisco. PCR products were subcloned into pCRII-TOPO (Life Tech.), ten clones were selected randomly, and sequenced by Core facility in MD Anderson Cancer Center.
Statistical analysis
Fisher’s exact test was used to examine associations between LEKTI expression and PNI and LV in human samples and clinical variables. Kaplan-Meier survival curves and log rank tests were used to examine the association between tumor expression of LEKTI and patient disease-specific survival. Multivariable analysis was performed using the Cox proportional hazards model. The data for this analysis were tested (plot of differences in log of cumulative hazard rates of test variable against time) and found to conform to the proportional hazards assumptions. Data are presented as mean±SD. Statistical comparisons between experimental groups were analyzed by unpaired Student’s t test, and a two-tailed p<0.05 was taken to indicate statistical significance. For analyzing the correlation, we used Pearson’s test and the p values are indicated. We also used Mann-Whitney rank sum test as indicated. All statistical tests are two-sided. Differences in variables that were not normally distributed were compared using a nonparametric test (Mann-Whitney U test). Again, a p value less than 0.05 is deemed statistically significant. All statistical tests are two-sided.
LEKTI expression correlates with perinuclear invasion (PNI) in patients with SCC of the tongue
We previously published that both LEKTI mRNA and LEKTI protein was clearly absent from the HNSCC lines UMSCC-1, OSC-19, Tu-138, JMAR, and DM14 compared to abundant message and protein in normal oral epithelial cells.48,50 To characterize the expression of LEKTI in primary tumors, we performed immunohistochemical analysis of paraffin-embedded, formalin-fixed specimens from 60 unselected patients with HNSCC from various primary sites and stages. During initial review of unselected HNSCC specimens for LEKTI expression by IHC, PNI was incidentally noted in 3 cases. In all three cases the tumors were negative for LEKTI expression, including tumor cells that invaded the perineural spaces. Photomicrographs from one of these cases showing a crescent pattern of PNI are presented (Figure 1 top panel). Although the correlation between LEKTI status and PNI invasion has not been systematically studied in the sample group, the incidental finding of reduced expression in tumor cells mediating PNI is consistent with the hypothesis generated from animal experiments in which LEKTI expression was correlated with reduced PNI.
Based on these anecdotal findings and our results from animal experiments, we investigated the relationship between LEKTI expression and PNI in a preliminary study using a more homogenous cohort of patients. The MDACC Tumor Registry was used to identify a cohort of 81 consecutive, previously untreated patients who underwent surgery as initial treatment for SCC of the oral tongue between 1990 and 1993. Expression of LEKTI in tumors from patients of this cohort was examined by IHC (using an automated stainer) and evaluated by two independent investigators, including a board-certified head and neck pathologist. LEKTI staining of tumors was evaluated as either negative, intermediate, or strong according to criteria described in methods section. A representative photomicrograph showing these three patterns are shown (Figure 1, bottom panel). When we analyzed the entire cohort for LEKTI expression we observed that LEKTI expression was negative in 38% (31/81), intermediate in 54% (44/81), and strong in 7.4% (6/81) of samples (Table 1). The distribution of staining patterns resembled that previously observed in unselected cases, demonstrating that the expression patterns can be reliably categorized.
The surgical pathology report of each patient from the cohort was then retrospectively reviewed for histopathological features of the primary tumor. The number of LEKTI tumors with each pattern of expression as a function of PNI status is also included in Table 1. PNI was present in 37% (i.e.12/31) of patients with LEKTI-negative tumors. Conversely, PNI was present in just 11% (i.e., 5/44) of patients with LEKTI-intermediate tumors and 16% (i.e. 1/6) of patients with tumors staining strongly for LEKTI. While a statistically significant difference in PNI status (P= 0.01, Fisher’s Exact test) was found between tumors negative for LEKTI and those with intermediate expression, the small number of patients in the LEKTI-strong group limited meaningful comparison (Table 1). Therefore, PNI was compared between patients with LEKTI-negative staining and patients with LEKTI-positive staining, whether intermediate or strong (Table 2). PNI was present in 37% (12/ 31) of patients with LEKTI-negative tumors. In contrast, PNI was present in just 12% (6/ 50) of patients with LEKTI-positive tumors. Therefore, in patients with LEKTI-negative tumors, the relative risk of PNI was 3.2 times greater (95% CI from 1.2 to 8.9) than patients with LEKTI-positive expression (p=0.007 by Chi Square Test).
LEKTI expression correlates with lymphovascular invasion (LVI) in patients with SCC of the tongue
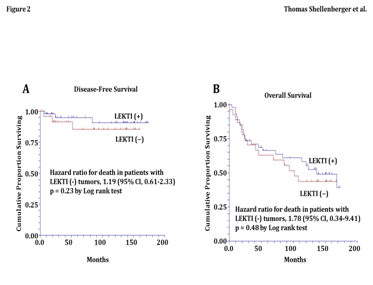
In our cohort of 81 patients, the patterns of LEKTI expression were also compared with LVI of the primary tumor specimen. LVI was present in 23% (7/30) of patients with LEKTI negative tumors (Table 3). In contrast, LVI was present in just 4% (2/51) of patients with LEKTI-intermediate or -strong tumors. Therefore, in patients with LEKTI negative tumors, the relative risk of LVI was 6.0 times greater (95% CI from 1.2 to 40.8) than patients with intermediate or strong LEKTI expression (p=0.01 by Fisher’s Exact Test). These data in patients confirm our findings in the orthotopic tongue xenografts and support the foundation of this proposal because they strongly support that the loss of LEKTI expression in SCC of the oral tongue correlates with a locally aggressive biologic behavior. Kaplan-Meier estimates of disease-free survival and overall survival showed that patients with LEKTI-negative expression had a 20% increased risk of disease recurrence (HR 1.2 and 95% CI 0.61 to 2.33, p=0.23 by Log rank test) (Figure 2A) and an 80% increased risk of death from all causes (HR 1.8 and 95% CI 0.34 to 9.41, p=0.48 by Log rank test) (Figure 2B). In contrast, analysis of the covariates for disease recurrence and death found no significant differences in age, gender, T-stage, grade, N-stage, and postoperative treatment between patients with LEKTI-negative and LEKTI-positive tumors (Table 4).
LOH is not a mechanism for loss of LEKTI mRNA and protein from HNSCC
In HNSCC biopsy specimens there appeared to be two patterns of LEKTI protein loss, with some tumors completely negative and others focally positive in areas where tumor cells were developing keratin pearls. Despite the fact that all established HNSCC tumor lines examined contained no LEKTI protein expression, some cell lines expressed mRNA. Together, these observations suggest multiple mechanisms for loss of LEKTI expression including both epigenetic and genetic abnormalities. The heterogeneous expression observed for LEKTI in tumor biopsies suggested an epigenetic mechanism of regulation. To further rule out the potential contribution of genetic loss, however, we analyzed 20 archival HNSCC biopsy specimens for LOH within the LEKTI locus using microsatellite markers found within the gene. Only 2/15 (i.e., 13.3%) of informative specimens had LOH within the LEKTI locus, making genetic deletion an unlikely mechanism.
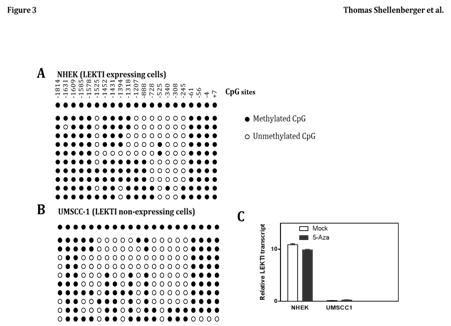
Promoter site methylation does not contribute to loss of LEKTI expression
Epigenetic change has been recognized as an important underlying mechanism of genetic alteration in human malignancies, including HNSCC. Therefore, we examined the methylation status of the LEKTI promoter region in UMSCC1 and NHEK cell lines. The promoter region of LEKTI expressing cells of NHEK was extensively methylated in several of the CpG sites (Figure 3A). Similarly, the promoter region of non-expressing cells of UMSCC-1 was also sporadically methylated at several CpG islands (Figure 3B). These results suggest no correlation between LEKTI expression and methylation of its promoter site. To establish that methylation was not responsible for silencing LEKTI gene expression, cell lines with LEKTI promoter methylation was treated with 5'-azacytidine, a demethylating agent. In each of the two cell lines tested, 5-azaC did not alter LEKTI expression as shown by real-time PCR (Figure 3C). These findings suggest that LEKTI expression is not regulated by promoter site methylation. We have not ruled out the mutational inactivation of LEKTI gene in HNSCC.
Transcriptional regulation accounts for loss of LEKTI mRNA and protein from HNSCC
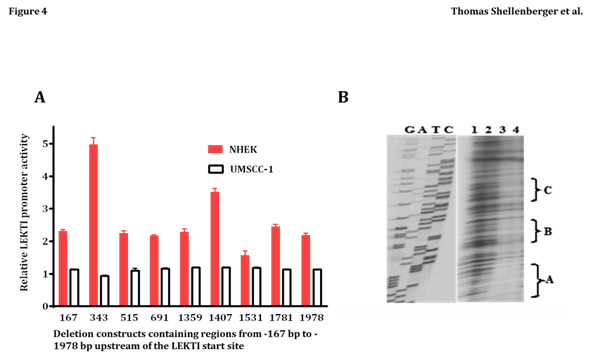
To begin addressing transcriptional mechanisms, we characterized the LEKTI promoter in primary cultures of NHEK. Primer extension analysis revealed a potential transcription start site (designated as +1bp) exactly 65bp upstream of the “ATG” start codon in exon 1. Inspection of the surrounding nucleotide sequence revealed a “TATAAAA” box 23bp upstream of the transcription start site. An identical transcriptional start site was predicted with the computer program Neural Network Promoter Prediction at: http://www.fruitfly.org/seq_tools/promoter.html. A series of deletion constructs containing regions from -167bp to -1978bp upstream of the LEKTI start site were cloned in front of the firefly luciferase reporter gene in pGL3, and transiently transfected into NHEK (which express LEKTI) or UMSCC-1 (which do not express LEKTI). As shown in Figure 4A, the 343bp and 1407bp constructs exhibited 5- and 3-fold greater activity relative to pGL3 alone in NHEK, but none of the constructs were efficiently transcribed in the LEKTI negative cell line UMSCC-1. Regulation in NHEK appears complex, as there seems to be repressor sequences contained between -343bp to -515bp, and between -1407bp and -1531bp. Also, there appears to be an enhancer sequence present between -1359bp and -1407.
In preliminary experiments designed to detect possible repressor sequences in the LEKTI promoter, a 312bp probe spanning regions -1235 to -1546 was incubated with UMSCC-1 nuclear extract in a DNase I protection assay. Increasing amounts of nuclear extract led to the protection of three distinct regions termed A, B, and C (Figure 4B, lanes 3 and 4). Sequences within these protected regions were found to overlap with several transcription factor-binding sites identified with the MatIspector program. In particular, protected region “C” overlaps with binding sites for two transcriptional repressor/activator proteins known as Barx2 and Engrailed-1, of which the former is abundantly expressed in HNSCC. Owing to the large size of the probe used in the DNase I protection assay, the probe actually overlapped sequences from the LEKTI promoter with both putative repressor and activator binding sites. Protected fragment A on the gel is actually found between -1382 to -1398bp, which occurs within the region expected to contain an enhancer (i.e., from 1359 to 1407bp). Interestingly, the protected sequence from “A” overlaps with a v-Jun recognition site. We have subcloned the 48bp fragment from 1359 to 1407bp containing the v-Jun recognition site and found that it indeed behaves as a transcriptional enhancer when cloned upstream of the thymidine kinase promoter and transfected into UMSCC-1 (data not shown). We have not tested the putative enhancer sequence transcription in NHEK but our results suggest that the sequence can activate transcription in UMSCC-1 in the absence of adjacent nucleotide sequences (which may recruit transcriptional repressors).
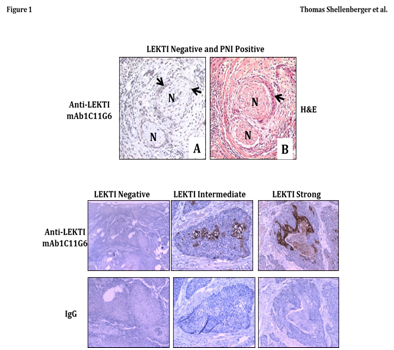
Figure 1 Characterization of LEKTI expression and PNI in tumor specimens from squamous cell carcinoma (SCC) of the tongue.
Top panel A tumor biopsy specimen from an unselected HNSCC specimen was examined for LEKTI expression by immunohistochemistry (A) and the corresponding region was also stained by H&E on a parallel section (B). In the first view, a crescent pattern of PNI is apparent (A), and LEKTI expression is absent from tumor cells invading the perineural space (A). Arrow heads point to tumor nests, and nerve segments are denoted with an “N”.
Bottom panel SCC tongue specimens were examined for LEKTI protein by IHC. Immunostaining was performed in with an automated staining machine (Dako) using an LSAB+ kit. Incubation of the primary antibody was performed with anti-LEKTI diluted at 1:150 for 60 minutes. Slides were analyzed by light microscopy. A typical example of each petern of LEKTI expression was shown. LEKTI expression was negative in 31, intermediate in 44, and strong in 6 patients.
LEKTI Expression |
PNI (+) |
PNI (-) |
Total |
Negative |
12 |
19 |
31 |
Intermediate |
5 |
39 |
44 |
Strong |
1 |
5 |
6 |
| Total | 18 |
63 |
81 |
Table 1 Pattern of LEKTI expression in PNI (+) and PNI (-) tumors from cohort
PNI is significantly different in LEKTI negative tumors compared to LEKTI intermediate tumors; P = 0.01 (Fisher's Exact test).
PNI in LEKTI negative compared to LEKTI strong; P =0.39 (Fisher’s Exact test).
LEKTI Expression |
PNI (+) |
PNI (-) |
Total |
Negative |
12 |
19 |
31 |
Strong |
6 |
44 |
50 |
| Total | 18 |
63 |
81 |
Table 2 PNI status of LEKTI positive and LEKTI negative tumors
P = 0.007 (Chi-square)
Relative risk (95% CI) of PNI in patients with LEKTI negative tumors compared to those with LEKTI positive tumors = 3.2 (1.2 to 8.9)
LEKTI Expression |
LVI (+) |
LVI(-) |
Total |
Negative |
7 |
23 |
31 |
Strong |
2 |
49 |
50 |
| Total | 9 |
72 |
81 |
Table 3 LVI status of LEKTI positive and LEKTI negative tumors
P = 0.01 (Fisher’s Exact test).
Relative risk (95% CI) of LVI in patients with LEKTI negative tumors compared to those with LEKTI positive tumors = 6.0 (1.2 to 40.8)
LEKTI (-) |
LEKTI (+) |
Comparison |
|
Age (years) |
54.8 |
54.4 |
p=0.87 by t test |
Gender ( percent male) |
40.0% |
32.2% |
p=0.64 by Chi Square |
T stage (%) T1 |
13% |
12% |
p>0.05 by Fisher’s Exact |
cN stage (%) cN0 |
22% |
43% |
p>0.05 by Fisher’s Exact |
Post op treatment (%) Sx |
57% |
69% |
p>0.05 by Fisher’s Exact |
Table 4 Analysis of the covariates for disease recurrence
These data confirm our previous in vitro and in vivo findings that the loss of LEKTI expression in HNSCC results in a cellular phenotype with locally aggressive behavior. Our findings shed new light on mechanisms of PNI and LVI, offer potential prognostic value, and may bring insight to patient selection for therapeutic approaches.
Funding
Supported in part by the NIH-NCI P50 CA097007, NIH R01 DE013954, NIH P30 CA016672, Alando J. Ballantyne Distinguished Chair in Head and Neck Surgery award, Michael A. O’Bannon Endowment for Cancer Research, NIH INRS Award T32 CA060374, and AAO-HNSF Percy Memorial Grant.
Author contributions
A.J, T.S, M.F, and G.L.C provided intellectual input into the design and presentation of the study. A.J, M.J, and T.S wrote the manuscript. Y.H, Y.K., and T.S carried out experiments. K.J. and R.J. organized data and reviwed literature. R.P. participated in project discussion.
The author declares no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.