MOJ
eISSN: 2374-6920


Oral mucosa keratinocytes are widely used in regenerative medicine. The unique cultured cell population “Epithelial-derived Pop-Up Keratinocytes (ePUKs)” was previously reported as undifferentiated cells. Gravity Assisted Cell Sorting (GACS) was used to isolate a small-sized population of undifferentiated cells enriched ePUKs.LC/MS/MS analysis was performed to define the cellular profile of ePUKs of primary human oral mucosa keratinocytes. Small sized ePUKs which showed increased expression of Dickkopf WNT signaling pathway inhibitor 1 (DKK1), serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 (SERPINE1), follistatin and tenascin-Crewe verified by Western blots. These proteins are involved in the regulation of cellular movement, hair follicle development and the maintenance of its stem cell niche. The fabrication of a tissue-engineered oral mucosa, ex vivo produced oral mucosa equivalent (EVPOME), using ePUKs showed increased abundance of these verified proteins. These findings indicate that the specific phenotype of ePUKs and their ability to influence wound healing promotion are implicated by highly expressed cellular movement regulatory proteins. Therefore, ePUKs may be a useful cell source for us in regenerative medicine.
Keywords: oral mucosa, quantitative proteomics, tissue engineering
EVPOMEs, ex vivo produced oral mucosa equivalents; DKK1, dickkopf wnt signaling pathway inhibitor 1; ePUKs, epithelial pop-up keratinocytes; GACS, gravity assisted cell sorting; FST, follistatin; SERPINE1, serpin peptidase inhibitor, clade e (nexin, plasminogen activator inhibitor type 1), member 1; TNC, tenascin-c
Oral mucosa keratinocytes are widely used for intraoral and extra oral clinical applications including reconstructions of cornea, urethral/bladder and esophagus.1 Rapid expansion of high quality cells is essential for decreasing the culture period resulting in prompt treatment and, mitigation of labor and cost. Therefore, a successfully isolated stem/progenitor cell would provide a robust source of cells for use in regenerative medicine. We defined “epithelial-derived Pop-Up Keratinocytes (ePUKs)”, which is human epithelial cells having high proliferative ability using a unique culture system and expansion technique.2,3 Further characterization of ePUKs is needed prior to their tissue engineering applications. ePUKs are floating, non-attached cells produced by large sized colonies in monolayer culture which are fed daily with 2-fold the usual amount of culture medium and the cell suspension containing ePUKs is poured into a new flask to form another monolayer without the use of enzymes to split the cultures.2 Small-sized cultured keratinocytes are reported as a progenitor/stem-cell-enriched population since they have a high ability of colony formation and long term proliferative capacity.4,5 However, when investigating the ePUKs undifferentiated profile, it is important to eliminate the contamination of the large sized dead or aged cells which are also floating in the media over laying a confluent mono layer, that lose their adherent ability. In our previous study, Gravity Assisted Cell Sorting (GACS) was reported as a unique method to separate cells by size.6 Isolation of small sized cells in ePUKs using this method provides us with an enriched undifferentiated small-sized cell population. We hypothesize that a comparison of selected small-sized cells in ePUKs versus traditional monolayer cultured cells would assist us in identifying specific markers, which would be useful to isolate them for clinical use. Thus, the aim of this study is to characterize small-sized ePUKs comprehensively by a proteomics-based approach to define their usefulness in regenerative medicine.
Procurement of human oral mucosa
Discarded keratinized oral mucosa was obtained from four patients; three males and one female with a mean age of 42.5years without any malignancies, undergoing tooth extraction and/orminor den to-alveolar surgery. The protocol for harvesting human oral mucosal tissue was approved by a University of Michigan Internal Review Board.
Primary oral keratinocytes and serial cultures and ePUKs culture
As described previously with modification,7 mucosal tissue was digested overnight with 0.04% trypsin solution (Life Technologies, Carlsbad, CA, USA) with 19.25µg/mL of gentamicin (Life Technologies) and 0.765µg/mL of fungi zone (Life Technologies) at room temperature, and transferred into 0.0125% trypsin-inhibitor (Life Technologies). Dissociated oral keratinocytes were re-suspended in a chemically defined culture system, complete Epilife (Life Technologies) and seeded into one T-25 flask. For serial cultures, cells were detached in 0.025% trypsin/EDTA (Life Technologies). For analysis, monolayer culture cells were fed adding 30mL of Epilife/150mm Petri culture dish every other day and collected after detachment in Enzyme Free Cell Dissociation Solution (Millipore, Billerica, MA, USA). ePUKs culture was previously described.2 The cells were fed adding 60mL of medium/ 150mm Petri culture dish, every 24hour. At confluence, the monolayer’s continued to proliferate; pushing keratinocytes into the overlying medium and the cells in suspension were collected as ePUKs. The monolayer cells underlying ePUKs culture were collected after detachment in Enzyme Free Cell Dissociation Solution (Millipore).
Gravity assisted cell sorting (GACS)
Gravity Assisted Cell Sorting (GACS) was performed as previously described.6 An ethanol sterilized funnel made from Millipore Nylon net filters (pore size; 11 and 20µm) was used to filter the cells tominimize size contamination in the small cell population. Each keratinocyte suspension was sized into two groups: cells trapped by a 20µm filter (large cells), and cells flowing through both a 20 and 11µm filter (small cells).
Protein lyses and digestion
Cell samples from one patient were lysed using RIPA buffer1 with proteinase inhibitor (Roche Diagnostics, Basel, Switzerland) and 10mM PMSF (Sigma-Aldrich, St Louis, MO, USA). Lysates were centrifuged at 15,000g for 10min at 4°C and supernatant was transferred to another tube. Protein concentration was measured by BCA method. 50µg of protein from each sample were reduced and alkylated in 100mm ammonium bicarbonate (pH 8) (Sigma-Aldrich) and 0.2% SDS (Sigma-Aldrich) solution using dithiothreitol (Sigma-Aldrich) and iodoacetamide (Sigma-Aldrich) reagent. The protein was precipitated by adding 5volumes of cold acetone and storing the sample overnight at -80°C. The resultant protein pellets were re-suspended in4M urea in 100mm NH4HCO3 solution. An enzyme mixture of trypsin-LysC (Promega, Madison, MI, USA) was added to each sample in a 1:50 (enzyme: sample) ratio and the samples were incubated overnight at 37°C.
Sample labeling and fractionation
The digested samples from the different cell populations were chemically labeled using amine-reactive tandem mass tags (TMT) (Thermo Fisher Scientific) following manufacturer’s instructions. The labeled samples were combined and desalted using C18 spin columns (PolyLC, Columbia, MD, USA). The sample was fractionated using high pH RPLC on a Thermo Electron Finnegan TSQ Quantum Ultra AM HPLC with an X Bridge C18 column (2.1mm i.d. x 100mm, 5µm particle size, Waters, Milford, MA, USA). Mobile phase A was 10mm ammonium formate in H2O (pH 10) and mobile phase B was 10mm ammonium formate in 10:90 H2O: acetonitrile (pH 10). A flow rate of 0.2mL/min was used with the following gradient program (time in min % B): 0min, 4%; 4.0, 4%; 5.0, 5%; 95.0, 50%; 110.0, 70%; 110.1, 90%; 115.0, 90%; 115.1, 4%; 120, 4%. Fractions were collected every minute and pooled together into twenty fractions by concatenated fractionation (e.g., fractions from 1, 21, 41, 61, 81, and 101minutes were combined).
LC/MS/MS
The samples were analyzed by LC/MS/MS on a Proxeon Easy-nLC II system (Thermo Fisher Scientific) and an Orbit rap Elite mass spectrometer (Thermo Fisher Scientific). Peptides were separated at a flow rate of 400NL/min on a 75μm i.d. x 25cm column packed in-house with Magic C18 AQ 100Å 5μm particle size material. A 130min linear gradient from 2 to 35% acetonitrile in 0.1% formic acid was used. The MS instrument was operated in positive ion mode. Survey MS scans (from m/z 400−2000) were acquired in the Orbit rap analyzer with resolution R=120000 at m/z 400, and the top 20 most intense ions were selected for tandem MS analysis by HCD. The normalized collision energy was set at 35% for MS/MS.
Data analysis
All acquired MS/MS spectra were searched against a concatenated forward-reverse database generated from the Swiss-Prot Human data base(downloaded April 2013) using the Andromeda search engine implemented in Marquand (v 1.3.0.5). Searches were performed using the following settings: precursor ion m/z tolerance: ±10ppm; MS/MS m/z tolerance: ±20ppm; up to two missed cleavages; static modification: carbamidomethylation (+57.02146 Da, C) and TMT 6-plex (+219.163 Da) of lysine sand peptide N-termini; dynamic modifications: oxidation (+15.99492 Da, M) and protein N-terminal acetylation (+42.011 Da). Identifications were filtered using a 1% peptide-level false discovery rate (FDR) and a 1% protein-level FDR. Quantification was performed using the intensity of TMT reporter ions.
Ingenuity pathway analysis (IPA)
In order to obtain detailed molecular information and infer significant signaling pathways from the proteome profiling results, differentially expressed proteins identified between sample ePUKs small vs. monolayer small were uploaded into the pathway analysis tool IPA (Ingenuity Systems, Redwood City, CA, USA) as previously described.8 The uploaded Excel file contains the relevant proteins with their fold change and corresponding primary accession number (Supplemental Table 4). The significance values for canonical pathways were calculated using the right tailed Fisher’s Exact Test by comparing the number of proteins that were involved in a given function or pathway relative to the total number of occurrences of these proteins in all functional/ pathway annotations stored in the Ingenuity Pathway Knowledge Base (IPKB).
Immunoblot
Cells from 3 individuals were lysed using RIPA buffer previously mentioned. For follistatin (FST), Dickkopf-related protein 1 (DKK1) and PAI-1 (SERPINE1), 50μg from monolayer small and 10μg from ePUKs small whole-cell extracts protein per lane were loaded and for tenascin-C (TNC), 50μg from each of the samples were resolved by SDS-PAGE and electrophoretically transferred to polyvinylidenedifluoride membranes. Membranes were then incubated overnight at 4°C with 1:200 anti-FST, (Sigma-Aldrich, HPA018155), 1:200 anti-DKK1,(Santa Cruz, Santa Cruz, CA, USA,sc22516), 1:500 anti-tenascin-C (Abcam, Cambridge, MA, USA, ab108930), 1:200 anti-PAI-1,(Santa Cruz, sc5297), or 1:1000 anti-β-actin (Cell Signaling Technology, Beverly, MA, USA, #4970) antibodies followed by 1:2000 goat-anti rabbit secondary antibody (Cell Signaling Technology, #7074) or horse-anti mouse secondary antibody ((Cell Signaling Technology, #7076). Signals were detected by the chemiluminescence reagent (Thermo Fisher Scientific).
Regenerative assay
Both ePUKs and monolayer cells used to manufacture tissue-engineered oral mucosa, EVPOMEs, were from the same cell strain. ePUKs were collected without centrifugation when monolayer cells reached at around 85% confluence. Monolayer cells were harvested by trypsinization and centrifugation. 1.9 x 105 cells of either ePUKs and monolayer cells were seeded onto 1cm diameter AlloDerm®s (Life Cell, Branchburg, NJ, USA) submerged in medium containing 0.06mM calcium for 24hours. On the following day, the medium was switched to growth medium containing 1.2mM calcium for four days followed by an additional seven days in air-liquid phase. Medium was changed every day. EVPOMEs were then fixed in 10% formalin.
Histological and Immuno Histochemical Characterization of Evpomes
Formalin-fixed, paraffin-embedded (FFPE) EVPOMEs were cut into 4.5μm sections, deparaffinized, and stained with hematoxylin-eosin for histological examination. For immunohistochemistry, FFPE-EVPOME sections underwent heat-induced antigen retrieval using a Low pH Flex flow pH 6.0 (Dako, Carpinteria, CA, USA) for 20minutes and were used after blocking endogenous peroxidase activity. The primary antibodies used were anti-FST 1:100, anti-DKK1 1:200, anti-PAI-1 1:200, anti-Tenascin-C1:100, for 2hours at room temperature. For control sections, the primary antibody was omitted. The sections were incubated with Flex Rabbit/Mouse envision system horseradish peroxidase- (HRP) labelled anti-rabbit polymer (Dako) for 90minutes and visualized with DAB (Liquid DAB+ Substrate SystemTM, Dako) for 2minutes at room temperature. Sections were counter-stained with Hematoxylin.
LC/MS/MS analysis of TMT-labeled peptides The study design is shown in the Graphical Abstract. Monolayer cultured (regular culture) keratinocytes, ePUKs and monolayer keratinocytes from ePUKs culture (double medium culture) were separated by size using GACS and the protein concentration of the different cell samples were determined, followed by reduction with dithiothreitol, alkylation with iodoacetamide, and digestion with a Lys-C/trypsin enzyme mixture. The digested samples were then reacted with amine-reactive TMT labels and mixed together before fractionation by high pH RPLC. The collected fractions were combined and analyzed by LC/MS/MS on an Orbit rap Elite mass spectrometer. A total of 53542 peptides and 11870 unique peptides were identified from samples analyzed on the LTQ-OrbitrapVelos. A final total of 6064 proteins were identified from samples by Max Quant. (A complete list of proteins identified in all the analyses is shown in (Supplemental Table 4).
Protein Profiles
The small-sized cells in each experimental condition which include the monolayer, ePUKs and monolayer from ePUKs culture contain an enriched population of undifferentiated cells while large-sized cells contain differentiated cells (Supplemental Table 1-5). 80% confluences of monolayer cultured cells are generally used in experiment or clinical therapy where once they reach 100% confluence, they are committed to growth arrest due to contact inhibition (Supplemental Table 4).3,9,10 Therefore, to elucidate a specific marker genuinely characterizing ePUKs, we compared serial monolayer small cells and ePUKs small-sized cells for further analysis. The differentially expressed molecular and protein functions in ePUKs small cells versus monolayer small cells involve cell death and survival, cellular growth and proliferation, nucleic acid metabolism, small molecule biochemistry and cellular movement (Table 1).
Name |
p-Values |
# of Molecules |
Cell Death and Survival |
6.88E-14- 1.74E-02 |
349 |
Cellular Growth and Proliferation |
3.10E-09- 8.69-03 |
340 |
Nucleic Acid Metabolism |
2.63E-08- 7.39E-03 |
65 |
Small Molecule Biochemistry |
2.63E-08- 1.05E-02 |
93 |
Cellular Movement |
1.06E-06- 1.44E-02 |
187 |
Table 1 Cellular and Molecular function of ePUKs Small vs Monolayer Small Population
Symbol |
Entrez Gene Name |
Fold Change Location |
Location |
Types |
DKK1 |
Dickkopf WNT signaling pathway inhibitor 1 |
14.64 |
Extracellular Space |
Growth Factor |
SERPINE |
Serpin peptidase inhibitor, clade E((nexin, plasminogen activator inhibitor type 1) |
12.849 |
Extracellular Space |
other |
FST |
Follistatin |
10.425 |
Extracellular Space |
other |
TNC |
tenascin |
4.328 |
Extracellular Space |
other |
Table 2 Differentially Expressed Proteins in ePUKs Small Population for Verification

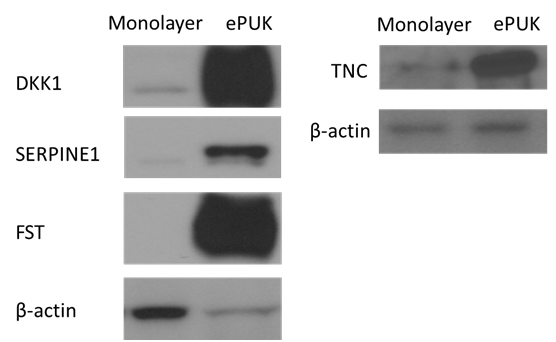

Verification of specifically higher expression in ePUKs small population by western blot and regenerative assay
We selected four proteins from the group that showed more than threefold difference between ePUKs small cells versus monolayer small cells and monolayer from ePUKs culture small cells from same cell strain (Table 2), (Supplemental Table 2 & 4). These proteins are all involved in cellular movement (Figure 1). DKK1, SERPINE1, FST, and TNC expression were validated by Western blot and intense expressions of those proteins were seen in ePUKs small cells (Figure 2). The regenerative ability of the monolayer cells and ePUKs cells was investigated by fabricating tissue-engineered oral mucosa, EVPOMEs. Both showed well stratified epithelium in H-E staining (Figure 3a & 3b). Immunohistochemical analysis revealed the increased expression of DKK1, SERPINE1 and follistatin in ePUKs EVPOMEs (Figure 3c-3h), while tenascin-C expression was similar between both cell populations (Figure 3i-3j).
EPUKs are grown with two major protocol changes: increased medium volume and more frequent medium changes, and were defined as undifferentiated cells in our previous study. The understanding of a cultured undifferentiated cell population might provide novel insights to tissue engineering where sustaining the cellular capacity of growth, proliferation and mobility is a key to the success of regenerative medicine.11 It has been reported that the majority of the ePUKs are a small-sized population,2 however, in culture supernatant, large-sized differentiated or dead cells are also floating where they have lost their adherent capacity secondary to their differentiation. Contamination of those large-sized cells cannot be ignored when we analyze the undifferentiated profiles. GACS was successfully used to enrich the population of small-sized undifferentiated cells.4 In our results, proteins having higher expression in ePUKs small cells are involved in cellular movement, indicating they might have enhanced ability for wound healing and tissue regeneration. Furthermore, immunohistochemical results showed that the increases in protein expression are seen in EVPOME device, which requires culturing 11days and induction of differentiation. Unlike other proteins, the intensity of tenascin-C expression in ePUKs after fabrication of EVPOME was weakened, indicating it might be affected by the change of culture conditions, which involves a differentiation process and being adherent to the scaffold. Application of ePUKs in regenerative medicine is suggested where expression of those molecules may be beneficial for wound healing after grafting. Further studies including in vivo transplantation are proposed for future tissue engineering use of ePUKs.
Given the present finding among the four proteins validated by western blot, DKK1 specifically inhibits the Wnt/beta-catenin signaling cascade to bind to low-density lipoprotein receptor-related protein (LRP) 5/6.12 Wnt-regulated developmental processes including posterior axial patterning, somitogenesis, angiogenesis, vasculogenesis, and organ formation are implicated in pathological events, including cancer and bone disease.12 It also regulates epithelial thickness and senescence in skin and oral mucosa.13,14 Follistatin is an antagonist of activin and a subset of TGF β super family molecules including myostatin and Bone Morphogenetic Proteins.15 Blocking activin action by pre-treatment with its binding protein, follistatin, modifies the inflammatory cytokine cascade, and reduces the severity of the subsequent inflammatory response and mortality.15 Limited activation of activin by follistatin in keratinocytes is beneficial for the wound healing process to prevent fibrosis.16 SERPINE-1 modulates detachment/re-adhesion cycles involving cellular migration through cell surface receptors including integrin and laminin.17 SEPINE-1 expression also correlates with tumor progression, where it is utilized as a cancer marker with poor prognosis.18–22 During epithelial would healing, SERPINE-1 is expressed at the wound edge where cell migration is important to achieve wound closure.17 Tenascin-C maintains the stem cell niche of the sub ventricular zone of the central nervous system, hematopoietic stem cell niches in bone marrow, corneal limbus and dental pulp.23 Regulation of cellular mobility and adherence to interact with fibronectin, integrins and heparin have an important role on wound healing.24–29 Interestingly, tenascin-C, DKK1 and follistatin orchestrate the hair follicle development and maintenance of its stem cell niche.30–32 High hierarchal progenitor population expressing these markers might be the reservoir of the high proliferative cells investigated in previous ePUKs study.2 As a similar concept of ePUKs, Chaffer et al.,33 identified that populations of human mammary epithelial cells cultured in their normal mammary epithelial growth medium contained a small proportion of cells that grew as floating cells above the majority population of differentiated adherent cells can revert to a undifferentiated state, indicating micro environmental signals to entering stem cell state including epithelial-mesenchymal transition may be provoked in those culture conditions.33 Another explanation could be cellular competition, which is reported as a phenomenon to exclude different phenotype of the cells both in vivo and in vitro, i.e., transformed cells in early stage of carcinogenesis or to coordinate the patterning and growth of normal tissues during development.34–36 In our present study, a heterogenic cell population of primary keratinocytes culture including differentiated cells, proliferating cells, or undifferentiated cells might cause cellular popping. Thus, under the environment of contact inhibition, differentiated keratinocytes may recognize undifferentiated cells as disparate neighbors and extrude undifferentiated cells from their society regulated by the specific signaling pathway, resulting in ePUKs.37
Cell death and survival are the most noted molecular and cellular functions in ePUKs small-sized population (Table 1). Most of these proteins are apoptotic induced proteins. Anoikis, the process of cell death, occurs in adherent culture cells when they lose their adherence should be considered here.38,39 The majority of the keratinocytes in suspension in ePUKs culture are routinely viable when transferred into a new culture flask, in the conditioned medium, to establish a new culture,2 indicating that even up to 24hours of floating in suspension does not affect their ability to adhere to a new flask surface and be proliferative. However, if the ePUKs cells in suspension are taken out from their original culture environment and re-suspended in fresh growth medium after centrifuging, their plating upon transfer and proliferation rate are lessened, indicating their fragility to changes in the culture environment and centrifuging.2 Induced apoptotic proteins in ePUKs small-sized population might be the cause of the mechanical damage seen in cells prior to processing before lysis or passing through the micro pore meshes and centrifuge. Methodological improvements should be considered in future experiments to minimize cell damage.
In conclusion, the comprehensive proteomics analysis of ePUKs small-sized population is successfully used to identify its specific molecular expression. Our studies suggest that the nutrient state of epithelial cells as well as other physical factors can modulate the expression of proteins possibly involved in the regulation of cellular movement. EVPOMEs made from ePUKs showed the increases in verified protein expression, indicating those are not temporarily expressed during floating status and still represented after induction of differentiation Application of ePUKs in regenerative medicine is suggested since expression of these molecules may be beneficial for wound healing after grafting. Besides the practical application to translational clinical therapies, the ePUKs primary culture can be used in studies to define the control of undifferentiated cells in epithelial tissue wound healing and development.
We thank Dr. Jianhui Zhuand Jintang He for their technical assistance and critical comments and Kristina Fields for her technical assistance of immunohistochemistry. We also acknowledge the National Institutes of Health under grant R01GM49500 (DML) and USA Department of Defense Grant to AFIRM I: W81XWH-08-2-0034 (SEF) for support of this work.
The author declares no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.