MOJ
eISSN: 2374-6920


The recent outbreak of the Ebola Virus Disease (EVD) urgently requires novel therapeutic approaches. While clinical trials are ongoing for an effective vaccine-based approach, a parallel effort to discover drugs is essential to provide a valuable alternative. Discovery of druggable targets for the EVD from the human proteome is a possibility. Using chemoinformatics approaches, the EVD associated proteins in the human genome can be verified rapidly for druggable structures. Using the Cancer Protein Annotation Tool from the CanSAR, a recently described 45 EVD-associated genes were analyzed for drug therapy use. Thirty-nine proteins were predicted to be druggable based on 3D structures and ligand binding potential. These proteins included a HIV associated chemokine, a coagulation factor, Heme enzyme and helicases involved in innate immune response to viruses, IFN-induced antiviral protein, an epithelial cell-specific transcriptional activator, a platelet factor and various members of the Human Killer cell immunoglobulin-like receptor family associated with fatal outcome with the Zaire variant of EVD. Ninteen of these proteins had 3D structural information available in the protein database including three enzymes. Based on ligand probability scores (>90%), three lead target proteins were identified (an enzyme, a blood factor and an epithelial cell specific transcription factor). Further, a lead drug-like compound (<1uM) was identified for the enzyme, Indoleamine 2,3-dioxygenase 1 (IDO1). A factor V antagonist was also identified in the study. The proteins described in the study offer a rationale for drug discovery approaches for EVD.
Keywords: chemogenomics, chemoinformatics, drotrecogin alfa (activated), druggable genes, ebola virus, human genome, ligand binding, protein 3d structures, sepsis
canSAR, integrated cancer drug discovery platform; CHEMBL, database of bioactive compounds at european bioinformatics institute, of the european molecular biology laboratory (EMBL); EVD, Ebola virus disease; KIR, killer cell immunoglobulin receptor; MAPK, mitogen activated protein kinase; ORF, open reading frames; PDB, protein database; RO5, rule of five
The current epidemic of the Ebola Virus Disease (EVD) in West Africa and its spread to different parts of the world is a major healthcare concern.1–6 The Ebola and Marburg viruses belong to the Filoviridae family of viruses and are responsible for causing highly lethal hemorrhagic fever.7–9 A US Centers for Disease Control and Prevention Report (Feb 15, 2015) stated that the death toll from the current EVD outbreak was 9,365 from a total of 23,218 cases. The incidence rate seems to be under decline for the first time since the outbreak. To date, there are no FDA approved drugs available for the treatment of EVD. An antibody-based therapy (ZMapp) was recently shown to be effective in a macaque model and in a limited number of patients.10 Several other experimental therapeutics are currently being explored involving nucleoside analogues, antivirals and antiretrovirals, antisense and siRNAs against the viral proteins, antibody cocktails and transfusion therapy.11–13 Clinical trials are underway to test the efficacy of the vaccines14,15 and if proven effective, they can become a strong deterrent for future spread of the disease.14 However, it is likely to take time and alternatives are needed to control this lethal disease.16,17
Repurposing currently approved drugs for the treatment of EVD can be of immediate benefit to Ebola patients. Three recent reports support this line of reasoning. Using an in vitro screen involving the Zaire Ebolavirus, FDA approved selective estrogen receptor modulators were identified which showed efficacy in mouse model of Ebola virus infection.18 In an another study involving an Ebola virus-like particle entry assay, FDA approved drugs were screened and the study identified 53 drugs which blocked the virus's entry.19 These drugs included microtubule inhibitors, estrogen receptor modulators, antihistamines, antipsychotics, pump/channel antagonists, antineoplastics and antibiotics.
In an another approach, using bioinformatics tools, 45 human proteins associated with EVD were identified; pathway mapping and drug bank screening of these genes identified antineoplastics, anti inflammatory drugs, estrogen receptor antagonists, leukotrienes, interferons, anticoagulants, nucleoside analogues, retinoic acid and statins.20 The 45 proteins identified in this study encompassed a chemokine, blood factors, interferon related genes, an epithelial cell specific transcription factor, heme and RNA helicase enzymes, a transporter and members of Killer cell Immunoglobulin Receptors (KIR). In addition, 16 previously uncharacterized protein Open Reading Frames (ORFs) emerged from this study.
Reasoning that these EVD-associated genes may offer novel target potential for the treatment of EVD, a chemoinformatics approach was undertaken to analyze them for druggableness. Results indicated drug therapy potential for 18 of these proteins including three enzymes. Ligand-based druggability analysis identified three putative lead targets, an enzyme, a blood factor and a transcription factor. A lead compound (<1uM) was identified for one of the enzyme targets. The protein targets evaluated in this study provide a framework for drug discovery efforts for EVD.
The bioinformatics and proteomics tools used in the study have been described.20–23 The protein annotation and chemical structure-based mining was performed using the canSAR 2.0 integrated knowledge-base, a publically available database.24 The browse canSAR section was used and the EVD-associated proteins were batch analyzed for protein annotations, 3D structures, compounds and bioactivity details. The protein 3D structure template models-related information was obtained from the Swiss Protein Database.25 The chemical structures were obtained from the CHEMBL.26 Comprehensive gene annotation for the EVD associated genes was established using the GeneCards,27 the DAVID functional annotation tool,28 the UCSC Ebola browser,29 and the UniProt30 databases. Protein expression was verified using the human protein Map,31 ProteomicsDB32 and the Multi Omics Protein Expression Database.33 The canSAR compounds link for genes has diverse filters such as activity and assay types, concentrations, molecular weight, RO5 violations, prediction of oral bioavailabilty and toxicophores. Putative drug hits were filtered from the canSAR datasets for the EVD-associated genes using Lipinski's rule of five (also known as Pfizer's rule of five), RO5. The RO5 is a rule of thumb to evaluate druggableness or to determine whether a compound with a certain pharmacological or biological activity possesses properties that would make it a likely orally active drug in humans.34–36 Highest stringency was chosen for the RO5 violation (value=0). Drugs with IC50 values, inhibitory activities and Ki values are chosen for the canSAR output. Toxicophore negative was chosen to filter the hits for toxicity associated compound structures.37
Chemoinformatics analysis of the EVD associated genes
To establish a drug therapy potential for the 45 EVD-associated proteins,20 the canSAR protein annotation tool was used for druggability prediction. Eighteen of these proteins were predicted to be druggable based on 3D structures with prediction (>90%) of druggable score (Table 1). These included blood factors Complement C1q (C1QB), Coagulation factor V (F5) and Platelet factor 4 variant (PF4V); C-C motif chemokine 8 (CCL8); probable ATP-dependent RNA helicases (DDX58, DHX58); Indoleamine 2,3-dioxygenase (IDO1); ETS homologous factor (EHF); Interferon-induced protein with tetratricopeptide repeats 2 (IFIT2) and Interferon regulatory factor 9 (IRF9); Pancreatic secretory granule membrane major glycoprotein (GP2); FXYD domain-containing ion transport regulator 3 (FXYD3) and ten KIR family members including a member of the Activating KIR family that is associated with Zaire EVD, KIR2DS1.38 Three of these protein hits had a very high probability for druggableness (>90%) judged by ligand-based druggablilty ranking (EHF, F5 and IDO1). Bioactivity results for two of the hit proteins (F5 and IDO1) identified an active lead compound for IDO1 (<1uM). The complete output from the canSAR analysis of the EVD associated genes is shown in Supplemental Table S1.
Ligand-based druggability of EVD associated proteins
In order to expand the backup capability for druggable targets for EVD, the ligand-based druggablity score was relaxed (>60% probability). This resulted in the identification of 14 genes with ligand binding potential. In addition to the top three hits (IDO1, F5 and EHF), several additional putatively druggable genes emerged including 1) members of the KIR family, 2) virus associated RNA helicases (DH58, DDX58), 3) a chemokine (CCL8) and 4) three ORF proteins (CXorf23, C2orf72 and C1orf200) (Figure 1 and Supplemental Table S2).
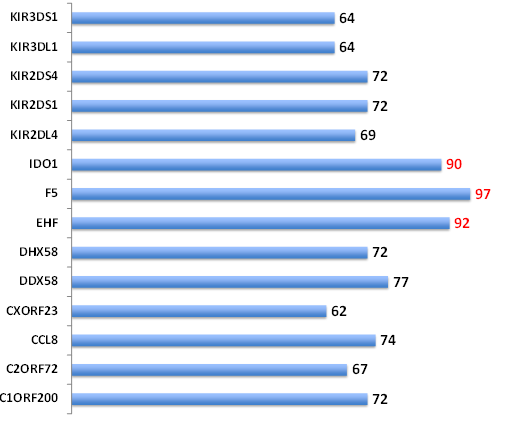
Figure 1 Druggability of the EVD-associated proteins. The canSAR protein annotation tool was batch analyzed using the list of 45 EVD-associated genes. Ligand-based druggability predictions are shown (>60% confidence).The % confidence is shown for each gene. Top three hits (>90% confidence) are indicated by red. KIR: Killer Cell immunoglobulin; IDO1: Indoleamine 2,3-dioxygenase; F5: Coagulation Factor V; EHF: ETS homologous factor; DHX58: Probable ATP-dependent RNA helicase DHX58; DDX58: Probable ATP-dependent RNA helicase DDX58; CCL8: C-C motif chemokine 8; ORF: Open Reading Frame.
Structural proteomics of the top three EVD-associated genes
Rational drug discovery approaches require molecular modeling and protein 3D structure information. Eighteen of the EVD associated proteins have 3D structure information available in the Protein Database (PDB). The 3D structures of the top three ligand-based druggable lead targets 1) Indoleamine 2,3-dioxygenase 1,39 2) Coagulation factor V40 and 3) ETS homologous factor41 are shown in Figure 2. The structures include alpha helices, beta strands and ligand binding sites.
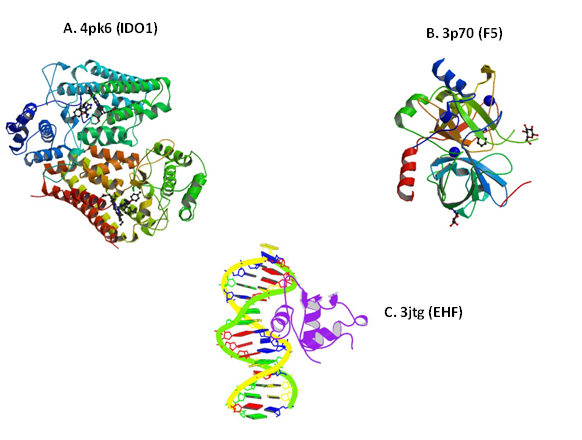
Figure 2 Structure of protein leads. The 3D structure of the top three EVD-associated proteins is shown. A) Crystal structure of the Indolamine 2,3- Dioxygenase 1 (IDO1) complexed with Imidazothiazole derivative; B) Five residues fragment of human Factor V, A2-B Domain linker is shown. Stoichiometry: Hetero 2-mer – AB and C) Crystal structure of mouse Elf3 C-terminal DNA-binding domain in complex with type II TGF-beta receptor promoter DNA, residues 289-391. Protein chains are colored from the N-terminal to the C-terminal using a rainbow (spectral) color gradient. PDB numbers are indicated. PDB: Protein Data Base; IDO1: Indoleamine 2,3-dioxygenase 1; F5: Coagulation Factor V; EHF: ETS Homologous Factor.
The IDO1 gene encodes indoleamine 2,3-dioxygenase (IDO), a heme enzyme that catalyzes the first and rate-limiting step in tryptophan catabolism to N-formyl-kynurenine. This enzyme acts on multiple tryptophan substrates including D-tryptophan, L-tryptophan, 5-hydroxy-tryptophan, tryptamine, and serotonin. This enzyme is thought to play a role in a variety of pathophysiological processes such as antimicrobial and antitumor defense, neuropathology, immunoregulation, and antioxidant activity.42 Through its expression in dendritic cells, monocytes and macrophages, this enzyme modulates T-cell behavior by its peri-cellular catabolization of the essential aminoacid tryptophan (NCBI and UniProt KB summary).
The F5 gene belongs to the multicopper oxidase family and encodes an essential cofactor of the blood coagulation cascade.43 This factor circulates in plasma, and is converted to the active form by the release of the activation peptide by thrombin during coagulation. It is a central regulator of hemostasis and serves as a critical cofactor for the prothrombinase activity of factor Xa that results in the activation of prothrombin to thrombin. Defects in this gene result in either an autosomal recessive hemorrhagic diathesis or an autosomal dominant form of thrombophilia, which is known as activated protein C resistance (NCBI and UniProt KB summary).
The EHF gene encodes a protein that belongs to an ETS transcription factor subfamily characterized by epithelial-specific expression (ESE). The encoded protein acts as a transcriptional repressor and may be involved in epithelial differentiation and carcinogenesis.44 The EHF gene acts as a repressor for a specific subset of ETS/AP-1-responsive genes and as a modulator of the nuclear response to mitogen-activated protein kinase signaling cascades (NCBI and UniProt KB summary). Additional details on these three lead proteins are shown in Supplemental Table S3.
Drug leads against EVD-associated genes
The canSAR drug bank has drug hits with bioactivity results for two of the EVD associated proteins, IDO1 and F5 (Figure 3). The IDO1 target had 535 compounds for IC 50 values in 77 binding assays in the CHEMBL library (target ID CHEMBL4685). Six hit compounds were identified with IC50 values <100nM. One lead compound for the IDO1 target was identified (CanSAR ID 489488; CHEMBL4685) which is active at 12nM (IC50) in bioassays for the IDO1 protein.45,46 This lead met the high stringency definition of putative drug (no toxicophore, RO5 violation: Zero, molecular weight <500). The chemical scaffold has 30 structures in the family thus allowing for chemoinformatics approaches for lead optimization efforts. The complete canSAR output for IDO1 is shown in Supplemental Table S4.
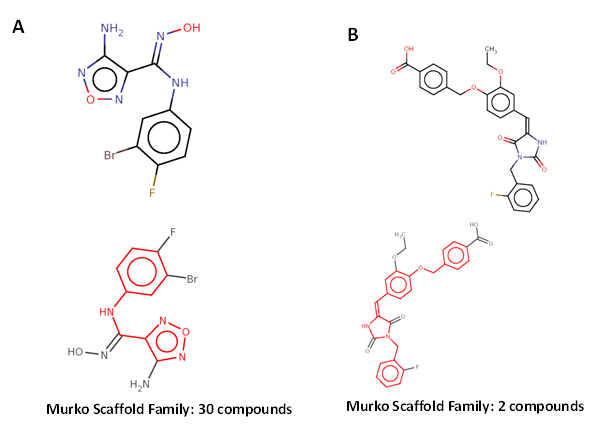
The F5 gene had 44 compounds for IC50 values in three cell-based assays in the CHEMBL library (target ID CHEMBL 3618). One putative drug-like hit compound was identified (CanSAR ID 438588, CHEMBL 259312) which is active at 2,500nM (IC50) in bioassays.47 This hit also met the high stringency definition of putative drug (no toxicophore, RO5 violation: Zero, molecular weight <500). The scaffold for this family has 2 structures, which can be used for lead optimization. The complete canSAR output for F5 is shown in Supplemental Table S5.
Currently no FDA approved drugs are available for the treatment of EVD. Therapeutic options for EVD include targeting the virus specific genes, use of neutralizing antibodies, vaccines and host specific genes whose functions may be required for the virus adsorption, entry or replication. Clinical trials for GlaxoSmithKline and New Link/Merck vaccines are underway in Liberia and additional trials are planned for Sierra Leone. The recent slowdown of the epidemic may however, affect the outcome of the trials. Further, the Ebola virus's acquisition of mutation(s) may compromise the efficacy of the vaccine. Thus, alternative therapeutic approaches including repurposing existing drugs are urgently needed.
The human genome is an attractive starting point for identifying host proteins for the therapy of diverse pathogens.48,49 HIV research provided a powerful example of discovery of the host cell-based target for therapy. The chemokine receptors (CCR5 and CXCR4) are the two major co receptors for HIV entry and have been successfully targeted for drug discovery.50 Among many CCR5 inhibitors developed so far, Maraviroc is the first drug that has been approved by the US FDA for the treatment of HIV. Hence, it was reasonable to develop a rationale in this study for gene target discovery for the therapy of EVD from the human genome. In a recent study, using bioinformatics approaches 45 proteins were identified using the Phenome to Genome and disease association tools.20 These genes provided a starting point for establishing a drug therapy potential.
Using the canSAR integrated protein annotation tool 14/45 proteins were predicted as druggable based on ligand binding scores. These 14 proteins included activating members of the KIR family, KIR2DS1 and KIR2DS3, both of which are shown to be strongly associated with the fatal outcome of the Zaire variant of EVD;38 a chemokine, CCL8, which showed a strong correlation with survival in Ebola virus infected nonhuman primates;51 RNA helicases; Coagulation Factor V; an epithelial cell-specific transcription factor, EHF; interferon regulatory factors; and IDO1 dioxygenase, a heme enzyme. In a recent study, these genes were also found to be differentially expressed in the nonhuman primate models with EVD, thus adding further credence to the EVD associated genes.51,52
Two of the three lead targets identified in this study have compounds identified in the canSAR database and offer a strong drug therapy potential for EVD and for other infections. Infection with the Ebola virus is accompanied by overexpression of the procoagulant tissue factor in primate monocytes and macrophages.8 These results raise the possibility that inhibition of the tissue-factor pathway could ameliorate the effects of Ebola haemorrhagic fever. In an Ebola infected primate model, inhibition of tissue factor-initiated blood coagulation prolonged survival time.53 Overexpression of tissue factor in Ebola virus-infected monocytes/macrophages resulted in fibrin deposits in spleen, liver and blood vessels of the infected macaques.8 Ebola hemorrhaging is due to formation of small clots throughout the blood vessels, which reduces the blood supply to organs. Thus an antagonist to Factor V could have a significant therapeutic benefit for EVD. The putative lead compound identified (canSAR ID 438588, CHEMBL 259312) offers a potential for further development as a Factor V antagonist. A Factor V inhibitor, a recombinant protein Drotrecogin alfa (activated) is already an FDA-approved drug for sepsis, and it shows efficacy in macaques infected with the Ebola virus.54 However, this drug was withdrawn due to failure to show a survival benefit55 and the putative lead identified in this study may provide an alternative. A drug-like inhibitor of the heme enzyme, IDO1 (canSAR ID 489488; CHEMBL 565923), emerged from these studies (IC50 12nM). This compound fits the classical definition of a putative lead (lack of toxicophore, bioactive at nM, RO5 violation, Zero). The IDO1 protein is associated with diverse bacterial and viral infections including Ebola, HIV, influenza, cytomegalovirus, hepatitis C virus, malaria and leprosy. Thus, an inhibitor of IOD1 may be beneficial for the therapy of these diseases. No compound was identified for the third lead gene, EHF, the epithelial-specific transcription factor. However, its association with EVD and key pathways such as Tumor Necrosis Factor Receptor (TNFR) and the Mitogen-Activated Protein Kinase (MAPK) pathways opens up novel opportunities for pathway-targeted drug discovery efforts.
At the present time evidence linking any of these proteins with Ebola virus binding or replication is, however, lacking. Nevertheless, in view of the urgent need for therapy of EVD, the targets and the lead drugs which emerged from this study should be tested in the animal models56,57 and in the in vitro screen.19
Currently limited knowledge exists about the host factors that are required for the Ebola virus. It is suggested that in epithelial cells, TIM-1(human hepatitis A virus cellular receptor 1) serves as a receptor for the virus.58 The cholesterol transporter Niemann Pick C1 (NPC1), present in the endosomal/lysosomal membranes, has also been shown to be required for productive infection.59,60 The genes identified in this study add to the growing list of putative host targets for the Ebola virus.
A random approach to drug discovery is a time consuming process with a very low probability of success. A rational molecular targets discovery by genetic association studies and the chemogenomics identification of bioactive, drug-like lead compounds can greatly increase the odds of finding the drugs for therapy. The leads can be rapidly tested in preclinical animal models for efficacy, bioavailability and toxicity. With the availability of chemical scaffold, the lead structures can be readily optimized. Such an approach can remove the randomness or the guesswork from the discovery process, thus saving valuable time.
Using bioinformatics and chemoinformatics approaches, druggable targets for EVD were established. The availability of protein 3D structures and ligand binding data for 18 EVD-associated proteins opens up new avenues for drug discovery efforts. The two lead compounds identified targeting Indoleamine 2,3-dioxygenase 1 and Coagulation Factor V can be tested for efficacy in animal models. The association of the EVD-associated target genes with multiple diseases suggests a broader use for these newly discovered compounds.
This work was supported in part by the Genomics of Cancer Fund, Florida Atlantic University Foundation. I thank the canSAR gene annotation tool for valuable datasets. I thank Jeanine Narayanan for editorial assistance.
The author declares no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.