MOJ
eISSN: 2374-6920


Understanding regulative potential and biological significance of miRNAs relies on our ability to correctly identify their target genes. Many experimental and computational efforts have been made to identify miRNA targets. However, reliable microRNA target prediction is still unsolved computational challenge. In this review we will focus on the basic principles and assumptions on which are based current computational approaches for finding and target prediction of miRNAs as well as the challenges for their future development.
Keywords: miRNA biogenesis, RNA folding, target prediction, sequence alignment
miRNA, microRNA; ncRNAs, non-coding RNAs; pre-miRNA, miRNA long single-stranded precursor; FEM, free energy minimisation
Encoded by eukaryotic nuclear DNA in plants, animals and some DNA based viruses miRNAs are a class of small ~21 nucleotides non-coding RNA (ncRNAs).1,2 Why these short starches of nucleotides are so important? Despite they small size it happens that miRNAs by binding to complementary sites on mRNAs gene transcripts are capable to induce cleavage or repression of productive translation for dozens or even hundreds of different mRNAs.3 As a result individual miRNAs can impact multiple cellular pathways like - signaling pathways; cell differentiation, proliferation/growth, mobility, and apoptosis; brain function, subcellular compartmentalization and chromatin remodeling.4–7 The ability to regulate almost every cell pathway makes miRNA very powerful factor in cells function and development. Understanding miRNAs will help in the discovery of treatments for diseases like cancer.8–10
Understanding regulative potential and biological significance of miRNAs relies on our ability to correctly identify their target genes. Many experimental and computational efforts have been made to identify miRNA targets.11–20 However, reliable microRNA target prediction is still unsolved computational challenge.21 In this review we will focus on the basic principles and assumptions on which are based current computational approaches for finding and target prediction of miRNAs as well as the challenges for their future development in time of incredible technological advances and fast growing number of sequenced genomes.
In animals miRNA gene is transcribed by RNA polymerase II in the cell nucleus in the form of long single-stranded precursor (pre-miRNA) transcripts that fold into very stable long hairpin structure. Starting from the cell nucleus and continuing on a few possible roads and processing steps toward the cytoplasm mature miRNA is excised from one of the stem arms of pre-miRNA hairpin. In the cytoplasm mature miRNA is loaded on a RISC protein complex and start to search for targets (Figure 1).22,23 The instruction for target recognitions is coded in a mature miRNA nucleotide sequence. Based on sequence homology, miRNAs can be grouped into sub-families, and many of them are evolutionarily conserved.24,25 Some miRNAs have tissue-specific or developmental-stage-specific expression patterns and their abundance can vary greatly depending on cell type.26
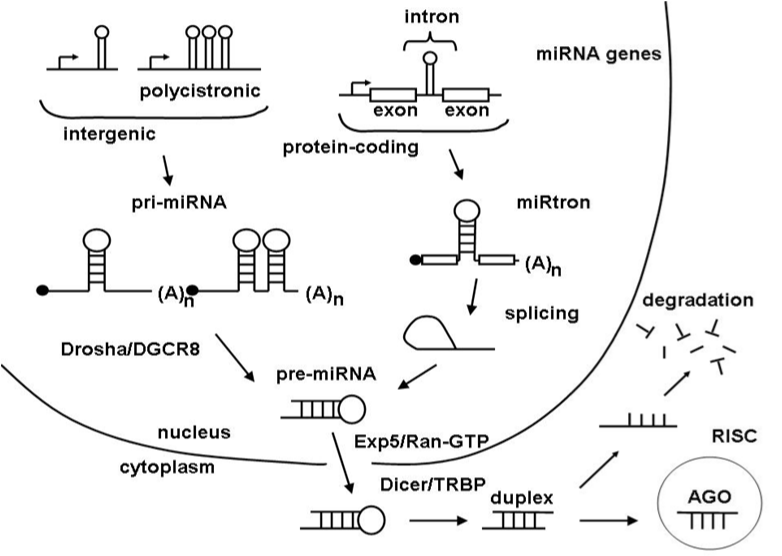
In plants, miRNA is transcribed and processed by homologous to the animals proteins, but the entire process of miRNA biogenesis is undertaken within the plant nucleus.27 The length of miRNA hairpin is more heterogeneous compare to animals, and can range from 70 to hundreds of nucleotides.28
A pre-miRNA is considered as an independent miRNA gene and seems to possess promoter and enhancer elements that are similar to those of protein-coding genes.29 MiRNA genes are scattered all over the genome but mostly they hide in intergenic regions or as part of introns of protein-coding and non-protein-coding genes in which case are excised by splicing machinery. MiRNA genes like to cluster together. Polycistronic transcripts with multiple miRNA-generating hairpins have been detected in animals but are rare in plants.30 Animal genomes have small but abundant miRNA families, while plants have fewer but larger miRNA gene families. Although many miRNA genes are conserved across species, the same gene family varies in size and genomic organization in different species.31,32 MiRNA community is dynamic – new genes and families are created and loss constantly.
There are a few major way miRNA genes can appear. First, the miRNAs genes in a given family have significant sequence homology to each other and almost identical mature miRNA sequences. This suggests that inside the families new miRNA genes appear by local duplications. MiRNA cluster formation by gene duplication has been observed in animals and probably dominates the evolution of plant.33 Second, there are indications that some miRNA genes in mammals resulted from transposons while convincing evidences are absent for plants.34 Finally, miRNA genes appear de novo from the many thousands RNA transcripts from non-coding DNA regions (more often from non-coding protein introns).35 Some of these RNA transcripts are expressed at low levels processed imprecisely and lack targets. Apparently they are subject to weak or no purifying selection and as a result easily form different secondary structures. It seems that these RNA transcripts form dynamical pool with high secondary structure plasticity for selection to act upon. It is proposed that in plants new miRNA genes emerge via mechanisms of inverted duplication events, while in animals new miRNA genes evolve gradually from intermediate non-functional hairpins structures.36 Once a functional hairpin structure is generated, it will start to interact with the miRNA processing machinery and different targets. At the beginning this interactions will be very week but gradually they will increase together with the expression levels of the new miRNA gene. This will give enough time for sequence-structure synchronization of miRNA transcript to adapt its sequence and kinetics of folding to provide specificity for every step of processing between transcription and integration of mature miRNA in particular biological pathway.
Technological advances such as bioinformatics and next-generation sequencing allowed the identification of a great number of miRNAs in different organisms in recent years. To date, over 2,000 human miRNAs have been identified and deposited in miRBase, while one to two hundred miRNAs are expressed in lower metazoans and plants.37
Given the increased number of sequenced whole genomes and the fact that much of these genomes are transcribed research on miRNAs especially finding and prediction of their target genes and the mechanism by which they repress their mRNA constitutes one of the frontiers in the study of post-transcriptional gene regulation.
miRNA transcripts pass through a few processing steps before mature miRNA is excised and loaded on the RISC complex. These processing steps require miRNA gen transcripts to have structures and sequences with certain biochemical characteristics. Therefore, finding of miRNA genes include discovery of pre-miRNA sequences which in equilibrium thermodynamic conditions can fold in known or new hairpin structure with certain characteristics that are important for proper biochemical processing.
RNA molecules at equilibrium have an ensemble of structures that they visit with different probabilities. How to find those with proper biochemical characteristics? Experimentally it has been shown that RNA transcripts of miRNA genes relative to other ncRNA transcripts and mRNA have unusual thermodynamically stable long hairpin structures.38–40 It seems that during the process of adaptation of structures and kinetics of folding of miRNA transcripts so that they could provide specificity for every step of processing between transcription and integration into particular biological pathway, a special set of sequences have been selected. As a result miRNA transcripts with selected sequences: have stable hairpin structures as their free energy minimum (fem) conformations; are separated by free energy gap from the other low free energy conformations; have conformations very different and separated with high free energy barriers from the fem conformation. This is not necessarily true for other ncRNA. For example, depending on their processing and biological functions, at thermodynamic equilibrium instead of one fem conformation (and structurally closed to the fem fluctuations) other ncRNA could be represented with different sets of inter converting secondary structures (switches). Sequences that can fulfill the thermodynamics requirements of these switches will be very different from sequences that fulfill requirements for fem conformations.
Therefore, we can formulate the problem of finding miRNA genes as the problem of finding the base-pairing that gives the lowest free energy change in going from the unfolded to folded state of their RNA transcript sequences. Also, we need to combine sequence, folding and structure considerations in a single integrated approach. Some additional filtering based on hairpin biochemical characteristics have to be done too like – sequence and structure requirements for Drosha and Dicer processing for example.41–43
Currently, RNA sequence dependence of free energy changes during the formation of secondary structures is not completely understood. Together with estimated 1.8n possible secondary structures for sequence with n nucleotides, this makes the finding of miRNA genes very difficult task.44 Nevertheless, dynamic programming algorithm, a type of recursive algorithm commonly used for optimization problems in biology, can search for polynomial time the entire set of possible RNA secondary structures and find the lowest free energy structure or particular sub-set from the ensemble of secondary structures available at equilibrium.45,46
Computational approaches can be divided in a few groups. In the first group one screen for sequences which can fold in a known experimentally verified pre-miRNA hairpin structure.17,47 The point here is that functional restrictions enforce more evolutionary pressure to preserve the structure than the sequence. So, the challenge is to minimize false positives. For example, the tested sequence can fold in a desire hairpin structure but is not the true miRNA sequence. Studies have shown that for pre-miRNA with stable thermodynamics hairpin structures the tested sequences nearly always have less favorable folding free energy compared to the true miRNA sequence even.17,39
In the second group one find a structure from a set of aligned sequences which is usually obtained based on similarity optimization.17,48,49 The point here is to identify sites showing correlated mutations. Such mutations can be interpreted as indication for functional or structural constraints. In other words one searches for mutations which preserve particular secondary structure.50
The assumption behind the above complementary approaches is that sequences with conserved catalytic activity, common evolutionary ancestry and having not enough time to diverge strongly between each other, fold into very similar structures.17,51 There exist a single optimal alignment for such set of sequences, which provides an accurate measure of similarity, structure, function and evolutionary history between them. However, with increasing evolutionary distances between nucleotide sequences, the single optimal alignment method is replaced by an ensemble of alignments of almost equal quality and ensemble of different folded conformations. Recurring difficulties associated with diverged sequence data include alternative alignment possibilities of insertions and deletions, region of length variations in which homology assessment is questionable or impossible, occurrence of localized excessive mutations to the point of saturation and lost of phylogenetic signals. Therefore, for diverged sequences optimizing similarity will not necessarily improve structure, function and evolutionary history assessments.52,53 This leads to false-positive miRNAs in the search procedures. Finally, in the last group one screen for sequences with new miRNAs. Here, the sequences or the structures of their close relatives are not known. In this case the false positives come from random sequences with no biological functions. It is widely accepted now that sequences which are biologically active are selected in such way that their biologically active conformations have more favorable free energies than random sequences.17,54
In the case of single sequence the free energy f of its fem conformation should be compare with free energy distribution of random sequences obtained from biological one by shuffling in such way that dinucleotide composition is preserved. The mean μ and standard deviation σ of fem conformations of a large number of shuffled random sequences is calculated. Statistical significance is expressed in standard deviations from the mean as Z-score . Negative Z-scores indicate that the tested sequence is more stable than the random sequences. However, single sequence methods are of limited statistical significance.
Some additional filtering based on hairpin characteristics have to be done too like – sequence and structure requirements for Drosha and Dicer processing; number and distribution of base pairs; length of the hairpin stem; location of miRNA on the hairpin stem; type of the bulges and their position on the hairpin stem; loop size and others.17
The most interesting and general case is connected with the so-called Sankoff’s problem for simultaneous optimization of alignment and folding.55 A simple reason behind the simultaneous optimization of alignment and folding is first synchronization of sequence and structure evolution and second that strong structural consensus among related, but diverged sequences are a good indicator for preserved functional role. Up to now there is no a general solution for this long standing problem. Whereas early methods focused on energy-directed folding of single sequences, now comparative analysis based on structure preserving changes of base pairs has been efficient in improving accuracy, and today this constitutes a key component in genomic screens. The usual approach is based on variations of - first fold than align and oppositely. For example, one can start with the sequences aligned by nucleotide identity and then find the conserved pairs in the given alignment or to predict low free energy structures for each sequence separately and then to sort through the predicted structures to find the structures common to all sequences.54 A recent approach for the general solution of Sankoff’s problem52,53 has shown that classical alignment algorithm and hybridization without intra-molecular base-pairing are mathematically equivalent. A simple analogy based on considerations of the frequency with which different nucleotides {A, C, G, T / U} or gaps occupy or not occupy certain positions in the alignment is shown on (Figure 2). The more substitutions of nucleotides or gaps are at a given position, the more often the formation of mismatches or loops are in these positions in the corresponding hybridization scenarios.
The equivalence between alignment and hybridization allows integrating both the energy-based and evolutionary-based information of sequence/structure covariations to predict the simultaneous folding/co-folding and alignment of RNA/DNA sequences. This can be done by integrating evolutionary and thermodynamic partition functions in the frame of dynamic programming algorithm (Figure 3).52
The regulatory information for target recognition and the degree of repression is encoded in the nucleotide sequence of miRNA genes. A key factor for inferring the regulatory information for target recognition is the analysis of miRNAs hybridization with mRNA of their target genes together with precise target-site localization.
The computational studies of the determinants of miRNA targeting specificity can be divided into two main classes: those that emphasize sequence features, and those that emphasize structural aspects.
Structural aspects include computational approaches for co-folding of two interacting nucleotide sequences including all possible conformational states like – matches, mismatches, bulges, symmetric and asymmetric interior loops, multi-branched loops and kissing loops in the regions where the two sequences fold separately and in the regions where they interact. Interaction regions and their conformational states are important for miRNA-mRNA interactions.56,57 Previous work revealed that 7-8 nucleotides of the so cold seed region at the 5’ end of the miRNA are responsible for target recognition.58 The seed region is small so is the target side, but it can be part of a bigger folded region of mRNA sequence. A portion of the mRNA where target site resides and is involved in intramolecular base-pairing has to become locally accessible to a targeting miRNA. The overall free energy to go from folded to unfolded state of this portion of mRNA and the hybridization of the target site of the unfolded portion with the 5’ seed region of miRNA is the driving force of this reaction. Therefore, the overall successes of miRNA binding to mRNA depend on mRNA length, its available secondary structures at thermodynamic equilibrium, binding side accessibility and thermodynamic stability of miRNA-mRNA duplex (Figure 4).56,59–61 One has also to include competition between different miRNAs or between miRNAs and proteins for the same target side. In this case concentrations of mRNA, miRNAs and proteins play crucial roll. The concentration of individual miRNAs in cells may vary by four orders of magnitude.62 This is even more important when there is more than one target side. Depending on the distance between target sides and miRNAs concentrations cooperative or competing mode of action have been reported to be predictive for the functionality of miRNA target sites. Effective modeling of saturation of binding and competition on binding sites is still in its infancy.63 Interestingly, recently it was found that miRNA genes that produce identical or nearly identical mature miRNAs have distinct biological activities that are controlled by their pri/pre-miRNA loop sequences. These raise the possibility of pri-/pre-miRNAs as target-recognizing RNA species.64 Sequence aspects include the sequence composition of the 3’ UTRs or of the immediate environment of the putative target sites to make interaction of target side with other portions of mRNA less effective and make target sides more accessible;59,65 the base-pairing pattern in the 3’ region of the miRNA;66 conservation of the miRNA binding site – it has been shown that conserved perfect seed matches to miRNAs have reduced SNP density which indicate that target sides are under purifying selection.67 It was also shown that nucleotide words complementary to miRNA seed regions are better conserved in 3’ UTRs than “random words” of the same length.17 However, there are experiments which question whether a seed match is either necessary or sufficient for miRNA repression: showing that perfect base pair matching does not guarantee interaction between miRNA and target gene, and wobble G:U base pairs are often tolerated in target sites.68 Taking advantage of miRNAs conservation we can use known mature miRNA sequence in one species to detect the probable location of its orthologue in another species, or further paralogues in the original species, by combining sequence analysis, sequence alignment and RNA secondary structure prediction.69 However, given the numerous rules and exceptions one needs an integrated statistical model to determine the most optimal prediction scenario.
While for finding miRNAs there is lot of attempts to approach the solution of Sankoff’s problem for simultaneous alignment and folding implemented in numerous software tools there are almost no attempts to consider simultaneous alignment and co-folding in the frame of comparative genomics. Co-folding is an integral part of our comparative approach.52,53 More importantly, concentrations of miRNAs, mRNA and proteins can be easily included too. This is very important for modeling the saturation of binding and competition on binding sites, and the extension of this approach is the focus of current research.
With almost 200047 reported miRNAs in human, and about 10 000 candidate genes, the network of interactions is extremely complex. Moreover, details about local concentrations and competitions among miRNAs in living cell are missing. Despite the advances in the recent years the mechanistic details of the function of miRNAs in repressing protein synthesis are still poorly understood70–72 and this reflects on quality of computational predictions. Thus, the degree of overlap between their lists of predicted targets is very poor and sometimes null.73
To improve computational miRNA target recognition approaches we need a deeper quantitative understanding of the miRNA-induced response and network regulation. The major problem with computational approaches is that it is difficult to predict, without sequencing data, when and in what type of cells a potential genomic hairpin will be expressed as RNA, and be available for processing.
This problem is address by the new technology RNA-seq.74 This technology enables not only the assessment of the expression level for a given miRNA loci, but also determines the exact mature sequence for that miRNA loci with a single base-pair resolution. This makes it possible to distinguish between miRNAs that share the same set of targets, but which can have small differences in their mature sequence.75
Based on RNA-seq It has been found also that:76
Some kinetic factors should be taken in consideration too. Such factors are transcription and turnover rate for RNA and protein molecules.77 Thus, the half-life of different proteins can vary from minutes to days whereas the degradation rate of mRNA would fall within 2-7hrs. Also, the rate of mRNA transcription is lower compared to protein translation in mammalian cells - 2 mRNAs transcribed per hour versus dozens of proteins per mRNA per hour.
Kinetic factors are important for cells which need to carry out a given function within a limited space of time. For such cells it is easier to find a direct correlation between the mRNA and protein levels than in less active cells. Therefore, when the aim is to find what genes are regulated by a given miRNA and how this explain a certain cell type function target predictions are necessary to guide target discovery. Moreover, connecting miRNA and their mRNA target genes to different biological response and functional pathways provides a unique opportunity to decipher the evolutionary forces involved in shaping miRNA network regulatory potentials.
None.
Authors declare that there is no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.