MOJ
eISSN: 2573-2919


Research Article Volume 5 Issue 2
1Institute of Molecular Biology and Biotechnology (IMBB), The University of Lahore, Pakistan
2University of Agriculture, Pakistan
Correspondence: Iftikhar Ali, Institute of Molecular Biology and Biotechnology (IMBB), The University of Lahore, Defence Road Campus, Lahore, Pakistan, Tel +92 345 4528975
Received: March 23, 2020 | Published: April 30, 2020
Citation: Shamim S, Mehboob S, Ali I, et al. Thermostable acidic lipase of Bacillus glycinifermentans-MK840989 isolated from contaminated environment; its optimization, purification and exploring potential applications. MOJ Eco Environ Sci. 2020;5(2):100-107. DOI: 10.15406/mojes.2020.05.00181
This study is the first report about isolation, purification and optimization of lipase from Bacillus glycinifermentans. In this study, Bacillus glycinifermentansMK-840989 was isolated from a local petrol pump. The bacterium showed lipolytic zones of 0.19cm, 0.044cm, and 0.28cm on peptone yeast agar, olive oil hydrolysis agar and chromogenic plate agar, respectively. B. glycinifermentans also produced an extracellular lipase (55.1µmol/ml). This bacterium preferred acidic environment (pH 5) for growing optimally at 80˚C when the medium was supplemented with 1% olive oil. The olive oil induced its growth up to 9h. The protein content of the purified lipase was estimated about 75mg/ml as compared to its crude form, i.e. 350mg/ml. The purified lipase was found to be thermostable acidic in nature as its optimum activity was observed at 90˚C (0.08U/ml) and pH 5 (0.02U/ml). Other optimization factors included 1% olive oil (0.065U/ml), 0.1mM maltose (0.023U/ml), 0.1mM Ca (0.025U/ml), 1% yeast extract (16.8U/ml), 1% wheat waste (0.019U/ml), 1% commercial detergent (0.016U/ml) and 1% tween-20 (0.015 U/ml). The purified lipase showed a polypeptide of 26.7kDa on SDS-PAGE. These features such as thermostability, acidic nature, ability to show activity in wheat waste and tolerance to detergents render the lipase of B.glycinifermentans MK-840989 as an attractive choice for biotechnologists to employ it at industrial level. The purified lipase of B.glycinifermentans MK-840989 can be a potential candidate for detergent and oil-remediation industry. It can help to replace conventional synthetic detergent as it is cost-effective and eco-friendly.
Keywords: Bacillus, environmental, enzymes, enzymology, optimization
Microbial enzymes are the biological catalysts that speed up the chemical reactions. They are cost-effective, eco-friendly and highly specific.1 The concept of enzyme is as old as human history.2 Almost 4000 enzymes exist, among which only 200 are in commercial use.1 Of them, 75% are of microbial origin.3 Microorganisms are known to produce many enzymes e.g. amylases, cellulases, proteases, etc.
Lipases (triacyglycerolacylhydrolases, EC 3.1.1.3) are serine hydrolases that convert triacylglycerides into glycerides (diglycerides or monoglycerides) by acting on carboxylic ester bond.4 They are extracellular enzymes and are induced by fatty acids, glycerol, oils, hydrolysable esters, etc.5 The production of lipase depends on bacterial strain, culture conditions and species.6 Different bacterial genera are known to produce lipases e.g. Staphylococcus, Bacillus,7 Pseudomonas,6 Burkholderia, etc.8–12
Pleigoet al.13 emphasized on the applications of enzymes at industrial level due to their number of applications in food, leather, dairy, detergents, flavor, pharmaceuticals, cosmetics, biofuels, etc.1 Moreover, the optimization of temperature, pH,5 agitation, substrate, etc. can enhance the enzymatic activity.14 The enzyme purification involves different steps such as ammonium sulfate precipitation, chromatographic techniques including hydrophobic interaction and ion exchange methods.15 Since high temperature directly influences the enzyme properties e.g. stability, reaction rate, solubility and lesser substrate viscosity as well as contamination, temperature-stable enzyme is desired.14,16 Moreover, the presence of metal ions is reported to improve its lipolytic activity.14,17
In this study, an effort is made to explore the lipase producing bacterium from indigenous environment. Here, we report the isolation of lipase producing bacterium, its characterization and growth optimization on different substrates. Furthermore, the lipase purification, optimal production and characterization were also studied.
Sample collection, isolation and purification of bacterial isolates
Different soil samples were collected from motor bike repair shop, oil change service at petrol pump, diesel at railway station, university car parking area and motor bike oil changing local shop. The isolation and purification of bacterial colonies was performed on Luria Bertani (LB) medium supplemented with 1% olive oil after sterilizing the medium. The cultural, morphological and biochemical characterization of the isolated bacterial colonies obtained were determined.18
Screening and selection of lipase producing bacteria
Following three assays were used to screen the lipase producing bacteria.19
Olive oil hydrolysis assay: In this method, 1% olive oil was added to LB agar plates (tryptone 10g, NaCl 5g, yeast extract 5g, and agar 15g) and zone of lipolysis was observed after incubating the plates at 37˚C for 24h.9
Peptone yeast agar assay: The peptone yeast agar medium was prepared by dissolving peptone 10g, yeast extract 10g, tween 80 3ml, olive oil 10ml and agar 15g in one liter of distilled water. The bacterial colonies were streaked on it and incubated for 24h at 37˚C. The zone of lipolysis was observed.
Chromogenic plate assay: A minimalmedium containing chromogenic substrate was prepared (0.01% phenol red, 4% tween 80, 20mM CaCl2 and 2% olive oil and pH 7). All the ingredients were mixed thoroughly in the form of a homogenized paste. Finally, 4% hot melted agar was added which made its appearance as orange-red color medium, followed by immediate pouring in the Petri plates and solidification at room temperature. By the help of a Pasteur pipette, wells were made in the agar medium and bacterial culture (overnight grown) supernatants added to the wells. The plates were incubated at 37˚C for 30min. The change in color from orange-red to yellow was the indication of lipase activity of the bacterial strains.20
Selection of efficient lipase producing bacteria
Based on the results of screening experiments as well as quantification assay, bacterium MK840989 was observed as maximum producer of lipase and thus selected for characterization.
Characterization and optimization of growth conditions of bacterium MK840989
The cultural, morphological and biochemical characterization of MK840989 was performed.18 The molecular characterization and identification on the basis of 16S rRNA gene was obtained from Macrogen® Korea. The optimum temperature, pH and carbon source for bacterium MK840989 was checked by the method of Liaqat et al.19
Optimum temperature: Different temperatures (25, 37, 50, 70 90˚C) were selected to check the optimum growth of bacterium MK840989 (Optical density at 580nm) after incubation for 24h at respective temperatures.
Optimum pH: The optimum pH was observed (OD at 580nm) by inoculating the LB broth media of different pH (4, 5, 6, 7, 8, 9) followed by incubation at 37˚C for 24h.
Optimum carbon source: Different oils (sunflower, canola, olive, mustard, mobile oil) were selected for this experiment. In 5ml LB broth, 1% of each of different oils (sunflower, canola, olive, mustard, mobile oil) was added as a carbon source followed by 1% inoculum. The test tubes were incubated at 37˚C for 24h. Next day, the optical density was observed at 580nm.
Growth curve analysis
The effect of oil on the growth of bacterium MK840989 was checked by inoculating the LB growth medium with 1% olive oil and 1% culture. The olive oil was not added in the control. The OD was read at 580nm after every 1h up to 9h.19
Quantitative analysis of lipase
Lipase quantity was determined by the method of Bussamaraet al.21 The LB medium inoculated with loopful culture and 1% olive oil was incubated overnight. Culture was harvested at maximum speed for 15min. In 100µl supernatant, 900µl solution A (1ml solution A: 1 ml isopropanol containing 3nol palmitate (pNPP)+9ml solution B: 200ml 50mM Tris-Cl pH 8.0, 0.2ml Tween 80, 0.8ml Triton X-100) were added. The mixture was vortexed properly and incubated at 37˚C for 30min. The increase in absorbance was read at 410nm. In control, the olive oil was not used. The lipase activity was calculated. One unit of lipase is the amount of enzyme liberating 1µmol of para-nitrophenol per ml per min under standard assay conditions.22
Isolation and purification of lipase
Lipase purification was done by the method of Liaqat et al.19 as described below.
Preparation of phosphate buffer 1M (pH 7.0): Stock solutions of 0.5M of Na2HPO4 and NaH2PO4 were prepared by dissolving 7.098g Na2HPO4 in 80ml distilled water (pH 7.0) and 5.99g NaH2PO4 in 80ml distilled water, (pH 7.0) respectively. The 1M phosphate buffer (pH 7.0)was prepared by mixing 30ml of 0.5M Na2HPO4 and 80ml 0.5M NaH2PO4. The final volume was made up to 1000ml by adding distilled water.23
Salt precipitation: In supernatant of 1ml fresh culture, 0.6g ammonium sulfate salt was added to obtain 90% purified enzyme. The salt was thoroughly homogenized by vortex for 10-15min. followed by cold incubation overnight. It was harvested at maximum speed for 15-30min. The supernatant of jelly-like consistency was shifted to a dialysis membrane. Before shifting, the Bradford assay was performed.24
Dialysis: A piece of dialysis membrane (5cm) was taken and its one end was folded and closed by a clamp. The jelly-like supernatant obtained in above step was pipetted from other end and closed by folding and clamped. It was suspended in a glass beaker containing 0.1M phosphate buffer pH 7.0 (prepared above) overnight. The beaker was placed in a refrigerator for overnight. The solution inside the dialysis tubing was shifted to an Eppendorf and centrifuged at following conditions; 1968 x g, for 10min, and at 4˚C. After harvesting, the supernatant was shifted to a new Eppendorf, and the Bradford assay performed. The sample at this stage was ready for column chromatography.
Colum chromatography: The column was prepared by placing a piece of cotton inside it followed by filling it with Sephadex G-100 (5g) and 2ml 0.1M phosphate buffer (pH 7.0). As Sephadex particles got settled inside the column, 100µl sample was injected to it. The sample was allowed to pass through the column and collected in collection tubes. The protein content of the collected fractions was determined by Bradford assay. The fractions showing more protein content were collected in one vessel. At this stage, the sample was ready for ion exchange chromatography.
Ion exchange chromatography: A column was filled with DEAE G-100. The sample collected in the above step was allowed to run in it. Similarly, the fractions of the sample showing high protein content were collected in one vessel. The Bradford assay was performed. The collected samples were lyophilized and stored at -20˚C. It was considered as a purified lipase which was used for optimization, SDS-PAGE and industrial applications.
Optimization of purified lipase
For optimization, 100µl purified lipase was used. The quantification of lipase was performed by the methods of Bussamaraet al.22 and Liaqat et al.19 at different conditions as follows. The activity of purified enzyme was checked over a wide range of temperature i.e.25, 37, 45, 50, 60, 70, 80 and 90˚C. Different pH (5, 6, 7, 8, 9) were selected to check the activity of a purified lipase. Moreover, the activity of lipase was checked with 0.1 mM Na, K, NH4, Zn, Cr, Ca, Fe, Mg and Mn ions. The 0.1mM of various carbon sources i.e. glucose, fructose, maltose, lactose and galactose were used as various carbon sources. Different oils (1% olive, sunflower, canola and mustard) were used. The wastes of banana, corn, wheat, sugarcane and pea peels were used as 1% in medium. The 1% peptone, yeast extract, casein and tryptone were checked. Finally, 0.1mM sodium dodecyl sulfate (SDS) was used in addition to some commercially available detergents (Ariel®, Bonus®, Bright® and Surf Excel®).The additives (1%) used were; tween 20, tween 80 and triton X-100.
Application assay for lipase
The potential application of the purified assay was performed as follows.9,19 One ml petrol was taken in a clean test tube and 1ml fresh supernatant was added and mixed thoroughly. It was allowed to settle down. The tube was observed for the formation of any layer. LB medium containing 1% kerosene oil and 1% petrol was prepared. It was incubated with the 1% bacterial isolate and incubated at 37˚C for 48h in shaking incubator. After 48h, the turbidity of the growth was observed. In order to perform detergent assay, three beakers were taken. The first beaker was filled with 50ml water, second with 50ml purified lipase and third with 50ml detergent of registered brand. Two clean pieces of white cloth were taken, one was marked with blue ink and ketchup was thrown on the other piece. Both pieces were allowed to dry at room temperature. They were cut in three equal parts and dipped in three beakers. They were placed in beakers for 1 hour followed by washing with tap water. After washing, they were air-dried and compared with each other. In Bio-surfactant assay on a clean glass slide, a drop of olive oil was placed. The sample (10µl) was placed in the center of the oil droplet. The change in the shape of oil drop was observed up to one minute.
Isolation and purification of bacterial isolates
Lipases are enzymes that are responsible for splitting triglycerides to diacylglycerides, monoglycerides and fatty acids.19 They hold special significance in various industries like food, cosmetics, pharmaceutical etc.4 A deep insight into their properties like growth conditions, inducers, inhibitors, interaction with metal ions, detergents, etc. should be taken into consideration.1 The microbial world known to produce lipase is diverse. For having practical application, always non-pathogenic strain is preferred. In this context, Bacillus stands out the list.19 In addition to its non-pathogenic nature; it is easy to culture as well. This study for the first time reports a novel strain Bacillus glycinifermentans-MK840989 as a lipase producer. For this study, mobil-oil contaminated soil samples were collected for the isolation of an efficient lipase producer. The most efficient lipase producer was Bacillus species which was observed to be B. glycinifermentans according to 16S rRNA sequencing. B. glycinifermentans was first time isolated from fermented soybean; it got its name from there.25 B. glycinifermentans are Gram positive, endospore-forming, motile and facultative anaerobes. To date, only three literatures related to B. glycinifermentans are published.25–27 In this study, it was isolated from oil change service from a local petrol pump.
Screening of lipase producing bacteria
All six bacterial isolates (SM-1, SM2 (MK840989), SM-3, SM-4, SM-5 and SM-6) were lipase producers. They showed following zones of lipolysis on olive oil hydrolysis agar plates: 0.021cm, 0.044cm, 0.031cm, 0.011cm, 0.029cm and 0.020cm by SM-1, SM-2, SM-3, SM-4, SM-5 and SM-6, respectively. Among all these, SM-2 (MK840989) showed more lipolytic activity. The same isolate showed more lipase activity (0.19cm) on peptone yeast agar plates as well as in chromogenic assay (0.28cm).
Lipase quantification and selection
Bacterial strain MK840989 showed the maximum amount of lipase (55.1µmol/ml). On the basis of results obtained from lipase screening methods as well as quantification, MK840989strain was chosen for further experiments as it was found to be a best lipase producer. It was identified as Bacillus according to the biochemical tests. The 16S rRNA sequencing showed it Bacillus glycinifermentans. Thestrain got registered in NCBI GenBank under accession number MK840989. It showed zone of lipolysis of 0.044cm on olive hydrolysis agar, 0.19cm on peptone yeast agar and 0.28cm on chromogenic plate. Its lipase was quantified and found to be 55.1µmol/ml. Liaqat et al.19 reported 42.7µM/ml lipase from B. stratosphericus-MK788130. In this study, the growth of B. glycinifermentans was observed optimum at 80˚C, pH 5 and 1% olive oil (Figure 1). Our results are in partial agreement with Liaqat et al.19 who reported the optimum growth of B. stratosphericus at pH 5 but at 50˚C. But it was in contradiction with Kim et al.25 who reported the optimum temperature range of B. glycinifermentans as 15-55˚C. Our results are in contradiction with the studies of Shivaji et al.28 and Ismail et al.29 who reported its optimum growth conditions of lipase producing Bacillus species as 8-37˚C, pH 6-10 and 34.8˚C, and pH 6.98, respectively. B. glycinifermentans-MK840989 cells were induced by olive oil (Figure 2) which agrees with Liaqat et al.19 but contradicts with the findings of Iqbal and Rehman,9 who observed slow growth of B. subtilis I-4 cells in the presence of olive oil. Adentunji and Olaniran,30 reported induction of B. aryabhattai SE3-PB lipase by using sunflower oil in the growth medium. The induction in the presence of oil gives a clue of its potential application in oil bioremediation.31
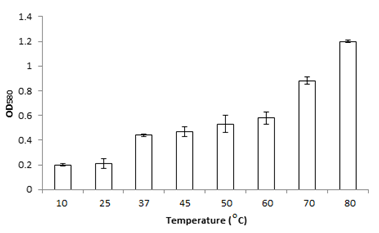
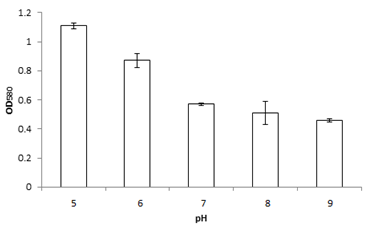
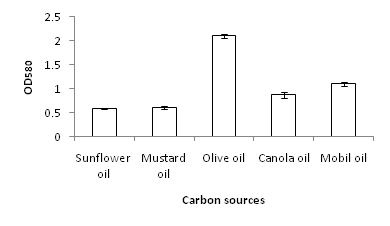
Figure 1 The optimum growth of B. glycinifermentans MK-840989 was observed at 80˚C, pH 5 and with 1% olive oil.
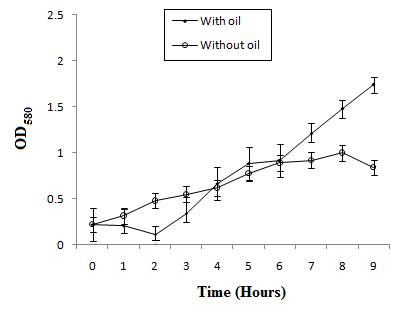
Figure 2 Growth curve study showed 1% olive oil as an inducer for B. glycinifermentans MK-840989 cells.
Protein content of isolated and purified lipase
According to Kornberg,32 the purification of enzyme from unnecessary impurities must be done as they hinder with its catalytic sites as well as its efficiency. The purification of bacterial lipases is previously reported by Andualema and Gessesse,33, Dey et al.,34, Rabbani et al.,35, Bhosale et al.,36, Sharma et al.,37, Liaqat et al.,19 In this study, the purification of B. glycinifermentans-MK840989 lipase resulted in decreased protein content from 350mg/ml to 75mg/ml (Table 1) which is in agreement with Borkar et al.38 who reported a similar decrease in protein concentration from 884 mg to 0.68 mg for P. aeruginosa SRT9 lipase during purification.
Enzyme purification steps |
Protein content (mg/ml) |
Supernatant (crude) |
350 |
After ammonium sulphate precipitation |
211 |
After dialysis |
98 |
After column chromatography |
88 |
After ion-exchange chromatography |
75 |
Table 1 Protein content (mg/ml) at different steps during lipase purification
Optimization of lipase activity
In this study,the purified lipase of B. glycinifermentans-MK840989 showed optimum activity at 90˚C, pH 5 and Ca+ (Figures 3a–3c), 0.1mM maltose, 1% olive oil, 1% yeast extract (Figures 4a–c), 1 % wheat waste, commercial detergent and tween-20 (Figures 5a–5c). Our results are in agreement with Liaqat et al.19 who reported the same optimum conditions (90˚C and pH 5) for lipase activity of B. stratosphericus-MK788130. According to Xiao et al.,39 lipase usually performs lipolysis between pH 4-11 and its optimum temperature ranges between 30-60˚C. According to Lomthaisong et al.,40 the lipase of P. xinjiangensis CFS14 showed maximum activity at 37˚C, pH 8 and in the presence of Mg2+ ions. Bacillus sp. W130-35 lipase, isolated from tidal mud flat showed its maximal activity at pH 9, 60˚C and Ca2+ ions.7,25 Tambekar et al.41 reported alkaline lipase production at pH 9 and 60˚C by B. flexus. In another study,42 the lipase of B. flexus was found functioning optimally at pH 10 and 70˚C. It was furthermore studied by the same group that this lipase was stable in tween-80 and triton X-100 which partially agrees with our findings (Figure 5c) as here maximum lipase activity was observed with tween-20 and minimum with triton X-100. B. flexus lipase also showed stability in the presence of commercially available detergents which strongly agree with our investigations.42 For B. subtilis I-4, the optimum enzyme activity conditions were; 50˚C, pH 7.0, Ca ions, olive oil and tween 80.9 Habibollahi and Salehzadeh,10 presented their investigations about lipase of Pseudomonas sp. KY 288051 as follows: 37˚C, pH 7, peptone and olive oil. The lipase of Staphylococcus aureus worked optimum (15.8U/ml) at 37˚C and pH 7 and peptone.43 In their study, olive oil induced the expression of lipase (12.5U/ml) which is in agreement with our results. Previous literatures also supported our results.44,45 The lipase obtained from B. cereus showed maximum activity at pH 8 (60.2 U/ml), 35˚C (55.25U/ml), maltose (66U/ml) and peptone (66U/ml).6 The lipase of B. sonorensis 4R had highest activity at 80˚C, pH 9, Mg and Ca ions.36 The lipase of a psychrophilic strain of Pseudomonas sp. LSK25 was obtained at 10˚C and pH 7.46 The purified enzymes isolated from Pseudomonas reinekei were found to be stable over a wide range of pH 5-9 and at 40˚C.47,48 According to Zarinviarsagh et al.,49 the yield of Ochrobactrum intermedium MZV101 lipase was 69% which was found stable at pH 10-13 and 70-90˚C. The fructose was found to be the best carbon source for optimal functioning of lipase (0.1U/ml) which contradicts with the findings of Sooch and Kauldhar,50 wherein it induced 53.2IU/ml lipase activity as compared to that by glucose (77.2IU/ml). In another study, 1.5% glucose was reported as the best carbon source for lipase activity (1590U/mg) followed by 1-2% fructose (1595U/mg) for Bacillus sp. ZR-5.51 According to Mazhar et al.,52 maltose made a maximum lipase expression (28.91U/ml) of B. cereus PCSIR NL-37 followed by fructose (27.2U/ml).53
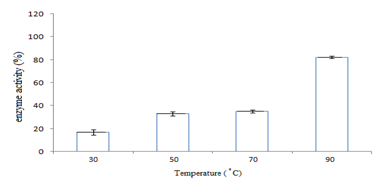
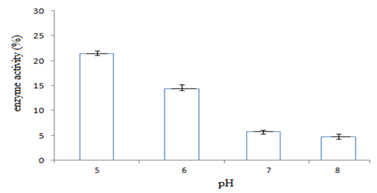
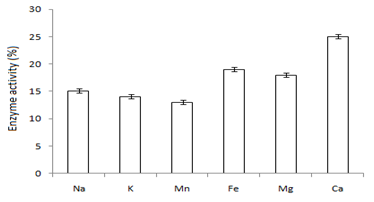
Figure 3 Graph showing optimum enzyme activity at different (a) temperatures (90˚C), (b) pHs (pH 5), and (c) metal ions (Ca+).
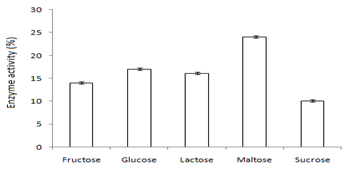
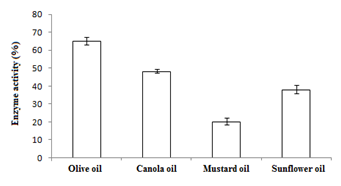
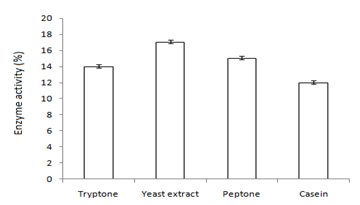
Figure 4 Graph showing optimum enzyme activity at (a) carbon sources (maltose), (b) oils (1% olive oil), and (c) nitrogen sources (yeast extract).
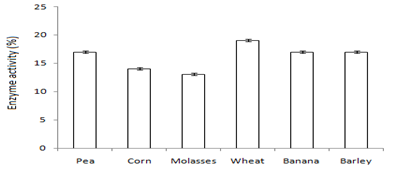
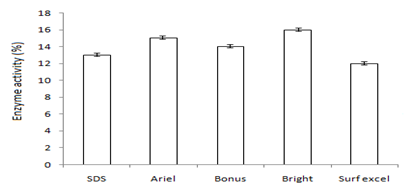
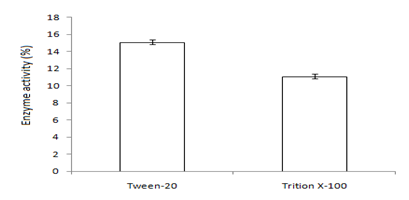
Figure 5 Graph showing optimum enzyme activity at (a) wastes (wheat), (b) detergents (Bright®) and (c) additives (Tween-20).
Potential industrial application
As the purified lipase of B.glycinifermentans-MK840989 was found to be thermostable and acidic, it showed a potential application as a detergent. The lipase of B. glycinifermentans-MK840989 showed potential application as a detergent. Our results are in contradiction with Liaqat et al.19 and Iqbal and Rehman,9 who reported the potential application of purified lipase of B. stratosphericus-MK788130 and B. cereus I-4 as a biosurfactant. Detergents are compounds that remove fatty acids and dirt particles.54 They are used in laundry for cleaning fabrics. They impart softness, anti-staticness and resiliency to fabrics. However, synthetic detergents persist for unlimited time period which contaminates the water bodies thus harmful for the environment. The green detergent or bio-detergents from microorganisms are the need of this hour.55 The previous literature already reported the property of microbial lipase as a detergent14,42 in Staphylococcus,56,57 Acinetobacter,58–61 Bacillus,15,20,62–64 Pseudomonas,65–67 Streptomyces68,69 and Burkholderia.59
Molecular weight of lipase
Here polypeptide of the purified lipase of B. glycinifermentans-MK840989 was of 26.7 kDa (Figure 6). Liaqat et al.19 reported 14kDa lipase from B. stratosphericus-MK788130. The lipase of O. intermedium MZV101 was of 99.42kDa as studied by Zarinviarsagh et al.49 Rabbani et al.35 reported 31kDa lipase from B. subtilis. Gururaj et al.70 observed lipase of 45kDa in Acinetobacter sp. AU07. Similarly previous literatures shows lipase of different molecular weights as 13.9kDa, 31.3kDa, 43kDa and 50kDa in psychrotrophic Pseudomonas ADT3,34 Enterobacter sp. Bn12,71 Staphylococcus SDMlip,72 Leuconostocmesenteroides subsp. mesenteroides ATCC 829373 and Pseudomonas reinekei,47 respectively.
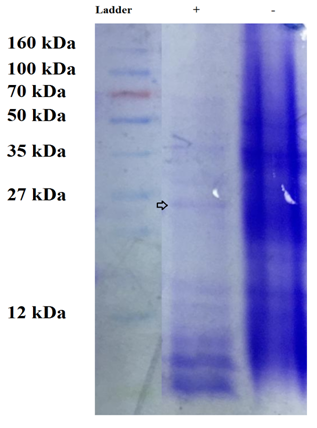
Figure 6 The SDS-PAGE showing the expression of lipase of 26.7kDa in the presence of 1% olive oil. The band of lipase is shown as encircled band in the figure.
Discussion
This study for the first time reported the isolation, purification and optimization of lipase from B. glycinifermentans MK-840989. It was found to be thermostable acidic bacterium which was induced by olive oil for the production of lipase. It possessed thermostable acidic lipase which enlightens its potential application in detergent and oil-bioremediation industries.
This study was not funded by any agency or organization.
None.
The authors declare no conflict of interest.

©2020 Shamim, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.