MOJ
eISSN: 2573-2919


After Mystacoleucus. Padangensis has been introduced to Toba Lake, North Sumatra, the status of M. padangensis stocks and their biological properties is unknown. In this study, we examine the genetic patterns of the M. padangensis in Toba Lake, North Sumatra Province. Employing the cytochrome c oxidase subunit I (COI) gene of the mtDNA, we profile the genetic variation within M. padangensis in Toba Lake. We also analyzed reproductive characteristic and commercial catches of M. padangensis. The result shown M. padangensis is a synonim for M. marginatus, in total, 1 polymorphic sites (represent singleton dimensions) was identified and phylogenetic reconstruction reveal low levels of genetic diversity with no clear pattern of haplotype–partitioning. Nucleotide diversity analysis infer the present of two lineages. The Ne value of M. padangensis (1,936 to 3,878), the population has not experienced population growth/expansion as expected and the total production of M. padangensis in Toba Lake of the year for 2013 has ranged between 3347,05 – 6694,1 ton. M. padangensis in the Toba Lake share similar life history traits include maximum body size, longevity, age at maturity, and fecundity (the number of eggs produced). M. padangensisis categorized as the opportunistic strategy consisted of fishes with short generation time, low batch fecundity, and low investment per offspring. This life history traits approach of M. padangensis are important for guide fisheries management for its sustainability. More over the information can be use as pilot data and apply it to data–poor species.
Keywords: M. padangensis, reproductive, DNA, toba lake, north sumatra
Mystacoleucus is a distinctive genus in the Cyprinid subfamily Barbinae, characterized by 6–10 branched rays in the anal fin, which is quite different from the other genus in this subfamily, whose anal fins normally have 5 branched rays.1 Mystacoleucus, has a wide distribution in Java, Borneo, Sumatra, Malaya, and Thailand2 and has seven species.3,4 Roesma et al.5 proposed that M. padangensis, is a new synonim for M. marginatus under the Indonesian common name wader. M. marginatus is a source of protein and of relatively high commercial value important for many local communities in the West Sumatra (Singkarak Lake). Reported price of M. marginatus has increased from IDR. 59.042/kg (2012) to over IDR. 80.000/kg in 2014.6 M. padangensis has been introduced to Toba Lake, North Sumatra intentionally to maximize their potential in 2003.7 There is no doubt that M. padangensis population in Toba Lake have experienced a very drastic decline since our last survey in 2013. However, limited information had been provided to understand the species’ ability to recover after declining to make informed conservation decisions as there is growing interest in this species in some small industry markets. Fishing is an important source of income and food for many peoples in Toba Lake. Good resource management is vital if continued economic progress and poverty alleviation is to be achieved. In many parts of Indonesia inland fisheries resources have become depleted due to inadequate management but, in the case of M. padangensis, there exists a current opportunity to ensure sustainable use. In this study, we examine the genetic patterns of the M. padangensis in Toba Lake, North Sumatra Province. Employing the cytochrome c oxidase subunit I (COI) gene of the mtDNA, we profile the genetic variation within M. padangensis in Toba Lake. We also analyzed reproductive characteristic and commercial catches of M. Padangensis. The results can provide important biological information as basic information, necessary to plan proper policy and management of M. padangensis.
Area study and sample collection
Sampling campaigns were conducted at six sites, Figure 1. and three times between April 2013 and November 2013 around Toba Lake, North Sumatra Province. Sampling and collecting representative samples of the adult fish community and fish juvenile (total sample size of 1078 individuals) were carried out using various types of fishing gear, Sulangat (a complex local fishing gear equipped with lamps), scoope net and gill nets.
Sample preservation and DNA analysis
Target species caught were identified to the species level following Kottelat et al.8 Immediately after fish was caught, tissue was taken from every individual and stored in absolute alcohol of 1.5 mL. In particular, at least 30 fishes collected for genetic study. Sampled fish were preserved in 10% buffered formaldehyde for several days after which the specimens were transferred to 70% ethanol. A voucher collection is stored at the Research Institute of Inland Fisheries, Palembang, South Sumatra, Indonesia. Juvenile fish at early life stages (from preflexion to postflexion larvae) were collected using two set of 30 cm diameter modified bongo nets during 15 min tows. The nets were maintained submerged for around 5 cm from surface. Larvae were kept in water and manually sorted after collection and were stored individually in absolute ethanol. Total genomic DNA was extracted from muscle tissue of each specimen using the Extraction Kit procedure ‘DNeasy Blood & Tissue’ (Geneaid). The partial fragment of mitochondrial Cytochrome C Oxidase Subunit–1gene (CO1) was amplified using modified universal primers described by Ivanova:9
Fish–COI–F (5'–ACT TCA AAC TTC CAY AAA GAY aty GG–3) and
COI–Fish–R (5'–TAG ACT TCT GGG TGG CCR AAR Aay CA–3').
Polymerase chain reaction (PCR) amplifications were made in a 50 µL of reaction volume consisted of 5 µL DNA samples, 16 µL double distillate water, 2 µL of each primer and 25 µL of PCR ready mixture solution (KAPPA). PCR cycling parameters included an initial denaturation phase at 95°C for 10 min, followed by 35 cycles at 94°C for 1 min, 48°C for 1 min and 72°C for 1.5 min and ended with a final extension at 72°C for 7 min. Finally, all amplicons were automatically sequenced in both directions at Macrogen S.A. Korea www.macrogen.com. The COI sequences of every analysed specimen were submitted to the GenBank database (MF062176–MF062186).
Chromatograms were controlled and checked manually edited with BioEdit (version 7.0.4.1)10 and multiple alignments were done using Clustal.11 Following alignments, COI sequences were translated to amino acids to check for the presence of premature stop codons that indicate the presence of nuclear pseudo genes or sequencing errors. Sequence divergence was estimated using the Kimura two parameters (K2P) model of base substitution.12 Phenetic reconstruction was done using a distance based method, Neighbor–Joining (NJ), carried out in MEGA5 software13 with the K2P model of substitution. Support for nodes in NJ analyses was assessed using non–parametric bootstrapping with 100 full heuristic pseudo–replicates. For comparative purposes, we used the several sequences of the freshwater fish species in Genbank (Accession Number view in Figure) to root the tree. We estimate of Ne to a benchmark value ostensibly important for conservation and management of M. padangensis in the Toba Lake Sumatra River. We also calculated the population history of M. padangensis and generated a mismatch distribution plot14 using DnaSP3.51 software.15 Detecting population expansion, past population growth or stability based on DNA sequences can be estimates employing a range of neutrality statistics15 such as Fu and Li’s D* test, Fu and Li’s F*, Fu’s FS test16 and Tajima’D test.17 All of calculations were computed using DnaSP 3.0 software.
Reproductive examination
In event level sampling, we go through our sampling frame selecting at least 30 individuals for each of sampling site. We measured the standard length (SL) and total length (TL) of collected M. padangensis to the nearest 1 mm, weighed the body (to nearest 0.01 g). The sex of each individual was identified and the gonads were assigned a developmental stage based upon morphology and macroscopic examination. Ovaries and testes were allocated a developmental stage according to the criteria modified Cassie18 Gonads from both sexes were subsequently preserved and stored in 10% buffered formaldehyde. Histological preparations were made from four ovaries and two testes representing each stage of development to check the accuracy of macroscopic staging and to better discriminate between mature and immature fish. Histological preparations were also made if a fish’s sex or developmental stage was uncertain. To prepare a gonad for histological examination, a small sample (<0.5 cm thick) was taken from the central portion of the preserved left gonad, embedded in paraffin, sectioned to 6 µm thickness, stained with Mayer’s haematoxylin and eosinY, and mounted on a microscope slide.19 All lengths reported here are standard lengths. Sex ratio, expressed as female: male, deviations from 1:1 null hypothesis were statistically tested by chi–squared test.
Total fish production assessment
The assessment should include a census in time on selected sites (six sampling sites) using enumerator (the enumerators examined the fish caught) and comprehensive interview and count number of active Sulangat, the primary means of capturing M. padangensis to determine the total fish production.
Genetic analysis
The PCR products of approximately 636–bp were obtained only from 11 M. padangensis Toba lake samples by amplification with Cytochrome Oxidase Subunit I (COI) universal primers, which spans sequence position 5537 – 6225 in GenBank HM 536913.1. In total, 1 polymorphic site (represent singleton dimensions) was identified in the sequence analysis of the 636–bp fragment among a total of 11 M. padangensis individuals from Toba Lake (Figure 2). Based on these polymorphic sites, two haplotypes were identified (Table 1), haplotype 1 was most frequent (90%) and the frequency of the remaining haplotypes was very low and resulted in a low nucleotide diversity of 0.00029 and low haplotype diversity 0.182. Variation of COI gene sequences of samples was summarized in Table 2. Based on our estimate of theta (θ) at 0.341 in M. padangensis, the effective female population size (Nef) was calculated to be 1,936 to 3,878. Given that effective population size inferred from the mitochondrial DNA is half of overall effective population size, and then we assume the effective population size (Ne) would be in between of 3,872 to 7,756.
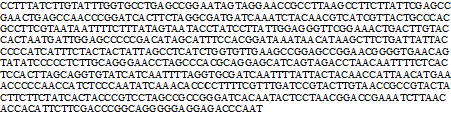
Figure 2 Nucleotide sequence of a partial DNA fragment (663 bp) of the M. padangensis COI gene. Single nucleotide polymorphism locations are designated in bold and underlined.
Haplotipe |
Polymorphic site |
|
|---|---|---|
5555 |
6054 |
|
haplotipe 1 |
T |
C |
haplotipe 2 |
T |
T |
Table 1 Position of bp variation of M. padangensis COI gene
Location |
S. size |
Sites plimrp |
trans |
Transv |
Indel |
Nucleotides composition |
Toba Lake |
11 |
1 |
1 |
0 |
0 |
T=27.4 |
C=28.3 |
||||||
A=26.7 |
||||||
|
|
|
|
|
|
G=17.6 |
Table 2 Location, sample size and genetic data of M. padangensis base on COI gene
Neighbor–Joining tree kimura 2 parameter (Figure 3) reconstructed from 15 sequences (11 sequences M. padangensis samples and 4 sequences samples as comparison). Base on phylogeny tree, common haplotypes of Toba Lake samples monophyletic with M. marginatus. Sequence analysis of the PCR products showed over 98% sequence identity to the COI gene of M. marginatus. The population history of M. marginatus population was examined using two different methods. A pair wise distance matrix was constructed with the 11 sequences of the COI of the mtDNA. The mismatch curve is bell–shaped (Figure 4), graphs observed mismatch distributions were compared with the expected ones. In the upper graphs expected curves were drawn by assuming constant population size whereas in the lower graphs expected curves were drawn under the assumption of population growth. The mean difference values for haplogroup Toba Lake was 0.182. Expansion results are presented in Table 3 Fu’s Fs test of neutrality was applied to further test the possible occurrence of population (Figure 3 & Figure 4).
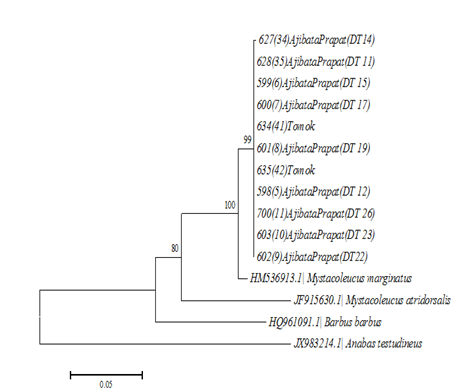
Figure 3 Neighbour-Joining phylogeny Kimura 2 parameter haplotype of M. marginatus base on COI gene.
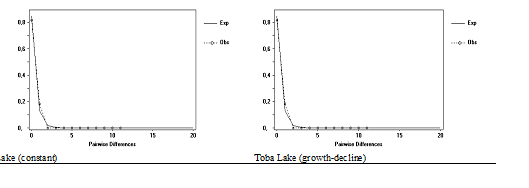
Figure 4 Observed and expected mismatch distributions for M. marginatus mtDNA COI sequences from Toba Lake under both constant and population expansion.
|
Toba lake |
sig |
Tajima's17 D |
–1,128 |
ns |
Fu et al.16 F* |
–1,289 |
ns |
Fu et al.16 D* |
–1,399 |
ns |
Fu et al.16 FS |
–0,400 |
|
Not significant P > 0,10 |
|
|
Table 3 Neutrality calculates for M. marginatus mtDNA COI sequences from Toba Lake. Not significant values in the data are indicated
Aspects of reproduction
Reproductive characteristics
padangensis make downstream migrations to reach very specific spawning locations in Toba Lake. Sex ratio was significantly different from the expected 1:1 ratio, (3,65:1), sex ratio shows a dominance of males in the Toba Lake (males/846 individuals, females/232 individuals during sampling periods). M. padangensis has an asynchronous ovary containing oocytes at various stages of development, Figure 5. Ovarian maturity was classified into the following four stages based on the most advanced oocytes contained in the ovary. Stage I oocytes correspond to previtellogenic oocytes and are characterized by a small size, a basophilic homogenous ooplasm, central or sub–central nucleoli and a high nucleoplasmic ratio. The developing phase (Stage II) includes oocytes in a primary to tertiary yolk stage and the mature phase (Stage III), ovaries much larger, occupying a significant part of the abdominal cavity.
Fecundity
Fecundity is the number of eggs in the female fish prior to release at the time of spawning fish. The fecundity is linked to longevity or weight of individual fish species.18 The result showed that the Bilih fish fecundity with total length between 87 mm – 136 mm with the number of eggs 4432–9887 ( 7125 ± 1753 ) (Table 4) (Figure 5). As for females, stage III testes, some spermatids remain in the lobule’s lumen, semen is never present within the sperm duct and even when squeezed vigorously, nothing comes out. Stage IV, almost similar with Stage III, but testes are characterized by lobules filled up with spermatozoa in the central part of the testes, semen is present within the sperm duct and when squeezed vigorously, sperm comes out (Figure 6).
Location |
N |
Matuirty |
Length (mm) |
Average (mm) |
Fecundity |
Egg diameter (mm) |
Panahantan |
15 |
III |
82–129 |
111.4 |
– |
– |
17 |
IV |
100–135 |
119.4 |
5557–9887 |
0.08–0.12 |
|
Ajibata |
10 |
III |
85–127 |
108.6 |
– |
– |
15 |
IV |
87–132 |
116.9 |
5459–8776 |
0.08–0.12 |
|
Tongging |
13 |
III |
85–129 |
10.5 |
– |
– |
16 |
IV |
101–131 |
116.1 |
4432–8166 |
0.08–0.12 |
|
Parapat |
13 |
III |
87–132 |
113 |
– |
– |
12 |
IV |
98–136 |
120.3 |
5541–8231 |
0.09–0.12 |
|
Paropo |
11 |
III |
87–132 |
108.9 |
– |
– |
11 |
IV |
89–129 |
116.8 |
4980–7851 |
0.09–0.12 |
Table 4 Fecundity and egg diameter of Mystacoleucus padangensis
Egg diameter
Egg diameter of M. padangensis range from 0.08 – 0.012 mm, it shown in Table 3. The results showed that the diameter of fish eggs is the same and the number of eggs that very considerably. The condition of maturity in level III and IV founded in every location in large number, It’s indicated that the Bilih fish will be spawning sustainable in Lake Toba.
Gonado somato index (GSI)
The highest male and female on maximum value of GSI Bilih fish found at the Tongging station (inlet of Lake Toba is located in Karo district) respectively at 12.23 and 12.79% (Table 5). The average value of the male gonad maturity index between observation stations based on Sapiro–Wilk test was not significantly different (a>0.05). The average value of GSI lowest male fish found in Paropo with value 2.77%.. The average value of GSI female found in Tongging with value 7.22%. GSI female values at each station were not significantly different (a>0.05). Based on Sapiro–Wilk test the average value of GSI between male and female fish are not significantly different observation stations. Based on the results of data analysis showed that the Variance of GSI are higher in male and female fish. The statement indicated that the fish can spawn almost all stations and sustainable spawn throughout the year.
Location |
Male/ Female |
N |
GSI (%) |
|||
|---|---|---|---|---|---|---|
Min |
Max |
Mean |
SD |
|||
Aji bata |
♂ |
40 |
1.27 |
12.17 |
5.18 |
2.89 |
♀ |
8 |
3.24 |
7.44 |
5.57 |
1.69 |
|
Panahatan |
♂ |
18 |
1.13 |
5.5 |
2.96 |
1.03 |
♀ |
18 |
1.25 |
12.51 |
5.82 |
3.67 |
|
Tongging |
♂ |
16 |
1.05 |
12.23 |
4.31 |
2.6 |
♀ |
5 |
3.42 |
12.79 |
7.22 |
3.92 |
|
Paropo |
♂ |
10 |
1.13 |
4.47 |
2.77 |
1.07 |
♀ |
12 |
3.46 |
8.41 |
5.57 |
1.32 |
|
Parapat |
♂ |
8 |
1.05 |
5.98 |
3.8 |
1.49 |
♀ |
9 |
3.77 |
6.33 |
5.43 |
0.9 |
|
Table 5 GSI of Mystacoleucus padangensis Toba Lake 2013
Total Production
Based on the results of studied of fishing gear (Ulangat) on Lake Toba found as many as 917 units (Table 6). It is founded in nearly all districts in Lake Toba. The data showed that the range of the catches fish (M. padangensis ) in Lake Toba by using Ulangat obtained by the range of 10–20 kg / day (Table 7). By converting the data is assumed that the potential of the bilih fish with ulangat amounted to 9.17 tons – 18.34 tons per day.
No |
Location |
Total of ulangat (Units) |
1 |
Aji Bata |
90 |
2 |
SiGaol–Gaol |
38 |
3 |
Simanindo |
59 |
4 |
Batu Gantung |
23 |
5 |
Penahotan |
17 |
6 |
Samosir |
107 |
7 |
Ambarita |
108 |
8 |
Onarunggu |
177 |
9 |
Nainggolan |
72 |
10 |
Palipi |
44 |
11 |
Pangururan |
182 |
Total |
|
917 |
Table 6 Total hand lift net (Ulangat) in Toba Lake 2013
No |
Location |
Local name of fishing gear |
Type of fish |
Cath composition (%) |
Total catch/Day (Kg) |
1 |
Aji Bata |
Ulangat |
Bilih |
100 |
10–20 kg |
2 |
Sigaol–gaol |
Ulangat |
Bilih |
100 |
15 –20 kg |
3 |
Simanindo |
Ulangat |
Bilih |
100 |
10 –20 kg |
4 |
Pangururan |
Ulangat |
Bilih |
100 |
10 –25 kg |
5 |
Onarunggu |
Ulangat |
Bilih |
100 |
10 –20 kg |
Table 7 Catch Compostion using Hand Lift Net in Toba Lake 2013
This study used nucleotide sequences from the cytochrome c oxidase subunit I (COI) gene region to examine the genetic properties of M. padangensis among six sampling sites in Toba Lake with a total of 11 individuals. Sequence analysis showed over 98% sequence identity to the COI gene of M. marginatus, this suggest that M. padangensis is a synonim for M. marginatus. Analyzed specimens were included to specific taxonomic group only if they formed monophyletic group with maximum 3% (K2P) sequences divergence.20 Using partial sequences of Cyochrome b sequence Roesma5 define that M. padangensis, is a new synonim for M. marginatus. The percentage of A+T base composition (54.1%) was much higher than C+G, which coincides with vertebrate protein–coding genes.21 Phylogenetic reconstruction of molecular variation between M. padangensis sampled from Toba Lake reveal low levels of genetic diversity with no clear pattern of haplotype–partitioning associated with a specific geographic location. However, when explored at the nucleotide diversity analysis infer the present of two lineages which is good for population longterm survival. Single–nucleotide polymorphism (SNPs) is important and now widely used for both, linkage analyses and biodiversity studies.22,23 A single, intermixing population of M. padangensis across the sampled location. No significant structuring was observed among other pairwise comparisons, this indicates gene flow between geographically adjacent locations. These results have direct management implications in recommending that M. padangensis from Toba Lake be managed as single stocks. In general, the active movement of adult and passive movement of eggs and larvae would accelerate the occurrence of gene flow among populations.24
It seems that the Ne value of M. padangensis (1,936 to 3,878) is sufficient to survive from a genetic viewpoint. The relatively high Nef reduces the probability of inbreeding among M. padangensis, potentially allowing the population to recover quickly.25 The population history of M. padangensis, suggest that the population has not experienced population growth/expansion as expected.26 More sensitive tests Fu’s Fs test of neutrality4 was applied to further test the possible occurrence of population expansion. However the Tajima’s D were positive in both populations and the index of Fu’s Fs test, was not significant at the 5% level for population for the pooled dataset. This is a weak indication of population expansion.27 To our knowledge, this is the first study to explore the genetic variation of M. padangensis in the Toba Lake to single stocks. Further, our data indicate M. padangensis at each sampling location likely had not undergone recent population expansion based on mismatch distribution analysis and neutrality tests. The same phylogeny means that M. padangensis in the Toba Lake share similar life history traits include maximum body size, longevity, age at maturity, and fecundity (the number of eggs produced). Referring to the organizing framework by28 which has evaluated patterns in fish life history traits across species, M. padangensis is categorized as the opportunistic strategy consisted of fishes with short generation time, low batch fecundity, and low investment per offspring. Each female fish has a finite amount of energy for reproduction. If all of this energy is used to create new fish biomass, then, at the extremes, it could be used to make one huge egg or billions of tiny eggs. Given these energy constraints, fish reproductive investment tends to be thought of as a spectrum of low investment in numerous offspring.29 This life history traits approach of M. padangensis are important for guide fisheries management for its sustainability. More over the information can be use as pilot data and apply it to data–poor species. It appears that the total production of M. padangensis in Toba Lake have been increased from 1,505 ton in 201030 to at least 3,347 ton in 2013. This data based on local gear used for fishing called Sulangat, a complex fishing gear which uses lights attached to structure above water to attract both fish and members of their food. In the every device, daily catches varied considerably between 10 – 20 ton and at least 917 Sulangat were installed in Toba Lake. Our study provides some information on the reproductive insight (histology) and total fish production of M. padangensis that would be helpful in similar studies and contributed to fisheries management of this species.31,32
None.
The authiors deckre there is no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.