MOJ
eISSN: 2573-2919


Rpp8 is present in Arabidopsis chromosomes in two different configurations: either as a tandem duplication with the loci separated by approximately 1kb, or as a single–copy gene. Polymorphism in gene copy numbers is common in clusters, and is also present in single–copy loci as the presence or absence of a locus. Copy number dynamics, intergenic exchange, and allelic diversity are all likely to be evolutionary responses to the same selective pressures for disease resistance. R–gene evolution, therefore, has both a vertical component across generations and a horizontal component throughout the genome, and each is likely to be shaped by natural selection for resistance. The presence of ancient and many segregating alleles at R–gene loci is interesting because disease resistance is thought to involve an evolutionary arms race between host and pathogen. Because of the lack of research on the polymorphism, this paper will investigate the polymorphism in each of the three loci, and determine how much of it is shared between the loci. From this, we can determine whether there is evidence for interlocus exchange, and if so, whether intergenic exchange is more common between the two members on the same chromosome (intra–chromosomal exchange), or whether exchange occurs more often between chromosomes. This paper also investigates whether these loci harbor more nucleotide polymorphism than that at another locus, which for the purpose of this lab can be considered to have "typical" levels of polymorphism for the species. From the neighbor–joined phylogeny, there is a significant amount of evidence for interlocus exchange. Around ~5% of the segregated DNA polymorphism is shared between loci and 95% is distinct to each locus. There is evidence for interlocus exchange. Intergenic exchange is more common between B and C loci than it is between any other loci combination. There is elevated polymorphism at Rpp8. All of these observations are coming from the hypothesis that the polymorphism demonstrated in this paper is coming from a continuously maintained balance of variation created by new mutations and natural selection. This seems to fit with the context of the gene sequences, as they are coming from Arabidopsis, which is self–fertilizing. Some future directions of research include looking for the cause of the increased polymorphism in Rpp8. More work should be done to clarify this relationship and compare it to similar relationships with other genes. Also more work should be done to find molecular evidence for this relationship.
Keywords: phylogenetics, morphology, polymorphism, evolution, biochemistry, bioinfomatics
The induction of plant defense responses by invading pathogens involves specific interactions between host and pathogen that bear important resemblances to human defenses against disease. These induced defenses mainly consist of multiple physiological responses, including localized cell death (called the hypersensitive response; HR), that result from the rapid phosphorylation of an array of proteins and the induction of genes through several pathways. Classical genetic studies of the HR, most notably for rust resistance in flax, have led to the gene–for–gene hypothesis, where a plant resistance (R) gene confers resistance specific to a corresponding avirulence (avr) gene in a pathogen.1 Overwhelming genetic evidence indicates that R–genes act as receptors for avr–gene products or avr–gene–dependent ligands, and effect changes in downstream gene action.2 In plant R–genes, polymorphism is often associated with loci that are present as tandem arrays of multiple copies. Due to promiscuous genetic exchange, paralogs within these clusters exhibit complex evolutionary relationships.3,4 Polymorphism in gene copy numbers is common in clusters, and is also present in single–copy loci as the presence or absence of a locus.5–7 Copy number dynamics, intergenic exchange, and allelic diversity are all likely to be evolutionary responses to the same selective pressures for disease resistance. R–gene evolution, therefore, has both a vertical component across generations and a horizontal component throughout the genome, and each is likely to be shaped by natural selection for resistance.8 The presence of ancient and many segregating alleles at R–gene loci is interesting because disease resistance is thought to involve an evolutionary arms race between host and pathogen.
A classic arms race is one that entails a series of selective sweeps as novel R–gene alleles, capable of recognizing pathogenecity determinants (called avirulence (Avr) factors) that previously avoided detection in a plant population, spread to high frequency.9 Support for these evolutionary dynamics centers on the common observation that amino acids evolve at a faster rate in functionally important regions of Rgene proteins than the corresponding rate of synonymous change.10 But according to the population genetics theory of selective sweeps, the rapid turnover of new R–gene specificity should cause a reduction in the age and number of alleles at a locus.11 Rpp8 is present in Arabidopsis chromosomes in two different configurations: either as a tandem duplication with the loci separated by approximately 1kb, or as a single–copy gene.12–14 The loci are labeled A, B, and C, respectively. Because of the lack of research on the polymorphism, this paper will investigate the polymorphism in each of the three loci, and determine how much of it is shared between the loci. From this, we can determine whether there is evidence for interlocus exchange, and if so, whether intergenic exchange is more common between the two members on the same chromosome (intra–chromosomal exchange), or whether exchange occurs more often between chromosomes. This paper also investigates whether these loci harbor more nucleotide polymorphism than that at another locus, which for the purpose of this lab can be considered to have "typical" levels of polymorphism for the species.15 This evidence will be used to look at the question of whether selection and/or frequent exchange has enhanced polymorphism in the R–genes.
Gene information
The focus of this paper will be on the Rpp8 gene. The Rpp8 data is contained in a file called "Rpp8_exon.nex"; it is a nexus file consisting of exon sequences for 6 A genes, 6 B genes, and 8 C genes from A. thaliana. There are also two sequences from one chromosome of a related species, A. lyrata, labeled Ce4B and Ce4A. The sequences are all aligned. The coding sequence is in frame.
PubMed Entrez
Entrez is a molecular biology database system that provides integrated access to nucleotide and protein sequence data, gene–centered and genomic mapping information, 3D structure data, PubMed MEDLINE, and more. The system is produced by the National Center for Biotechnology Information (NCBI) and is available via the Internet. Entrez covers over 20 databases including the complete protein sequence data from PIR–International, PRF, Swiss–Prot, and PDB and nucleotide sequence data from Gen Bank that includes information from EMBL and DDBJ. The Entrez retrieval system uses an intuitive user interface for rapidly searching sequence and bibliographic data. A unique feature of the system is its use of precomputed similarity searches for each record to create links to "neighbors" or related records in other Entrez databases. These links facilitate integrated access across the various databases. An Entrez global query provides search capability for a subset of Entrez databases at one time. Results may be viewed in various formats including Flat File, FASTA, XML, and others. A graphical interface provides easy visualization of complete genomes or chromosomes, as well as biological annotation on individual sequences. Entrez also allows Batch downloads of large search results. This was used to find sequences of ferulate–5–hydroxylase (FAH1) and A. lyrata (L.) O’Kaneb & Al–Shehbaz.
HyPhy
HyPhy (Hypothesis Testing using Phylogenies) is an open–source software package for the analysis of genetic sequences (in particular the inference of natural selection) using techniques in phylogenetics, molecular evolution, and machine learning.1 The paper uses this software to compare the independent and dependent phylogenies through the use of a bootstrap analysis. The bootstrap was run within normal and default parameters. In the bootstrap, the minimum number of simulation recommended was 100. The program allows 100–1000 simulations. For this analysis, 550 simulations were run. The simulations calculated the LR value. The LR value is the likelihood ratio, defined as 2 (log L− log L0), where LA is the likelihood for the alternative hypothesis, L0 is the likelihood for the null hypothesis (refer to the documentation for HyPhy).1 A simulation in the bootstrap is to pick random sites from the original sequence with replacement, rebuild the phylogenetic tree for two hypotheses, calculate the log likelihood and generate one likelihood ratio. The goal is to see the likelihood ratio from the data fall into the empirical distribution. In the program, the null hypothesis was entered and the alternative hypothesis.1 A null hypothesis supports the hypothesis that there is no significant difference between specified population; that any observed difference being due to sampling or experimental error. The alternative hypothesis supports the hypothesis that there is a significant difference between specified populations and that these differences share a cause. If the p–value is really small, ~p<0.0005, the null hypothesis is rejected. The possibility of proximal and distal genes evolving independently as the alternative hypothesis. And the possibility of proximal and distal genes evolving independently as the null hypothesis.1 The MEGA software was also used to align and organize the DNA before the phylogenies were created.
DNASP
DnaSP (DNA Sequence Polymorphism) is a software package for the analysis of DNA polymorphisms using data from a single locus (a multiple sequence aligned –MSA data), or from several loci (a Multiple–MSA data, such as formats generated by some assembler RAD–seq software). DnaSP can estimate several measures of DNA sequence variation within and between populations in noncoding, synonymous or nonsynonymous sites, or in various sorts of codon positions), as well as linkage disequilibrium, recombination, gene flow and gene conversion parameters. Moreover, DnaSP can conduct a number of neutrality tests, such as (among others), the Hudson, Kreitman and Aguadé (1987), Tajima (1989), McDonald and Kreitman (1991), Fu and Li (1993), and Fu (1997), Ramos–Onsins and Rozas, Achaz (2009) tests, and compute their confidence intervals by the coalescent. The results of the analyses are displayed on tabular and graphic form. This was used in the paper in order to calculate the Fisher and G–tests results.
MEGA
MEGA (Molecular Evolutionary Genetics Analysis) is software that specializes in analyzing FASTA DNA sequences. The software emphasizes the integration of sequence acquisition with evolutionary analysis. It contains an array of input data and multiple results explorers for visual representation; the handling and editing of sequence data, sequence alignments, inferred phylogenetic trees; and estimated evolutionary distances.2 The software allows the user the ability to browse, edit, summarize, export, and generate publication–quality captions for their results. MEGA also includes distance matrix and phylogeny explorers as well as advanced graphical modules for the visual representation of input data and output results. The main features of this software used in this paper are the phylogeny construction software and the substitution software.2 The substitution software will analyze the DNA sequences that are uploaded onto the program, after which, it will calculate the AiC value for each substitution model. The model with the lowest AiC value will be the model that will be used to analyze that sequence. See the supplementary information for more specifics on the AiC calculation. After the substitution model was determined, a phylogeny would be created with the gathered information. All of the phylogenies created using the Neighbor–Joining method.2
To begin the analysis, a phylogeny was created using the Rpp8 sequences (Table 1). This was a simple phylogeny that was created with neighbor–joining techniques (Figure 1). From the phylogeny, it is demonstrated that there is interlocus exchange between the A, B, and C loci. In the phylogeny, it is shown that there is significant overlap. This overlap is shown by the general shape of the phylogeny. Each clade of the phylogeny is directly connected to each other clade. In the phylogeny, there is no true outlier group. Every branch shares significant similarities with other branches within and in different clades. Also, the phylogeny is in two clades. There is one “outlier,” WUA that shares remarkable similarities with the rest of the gene sequences. And the second clade is one big clade that devolves into smaller clades. This de–evolution of the clade design shows significant amount of genetic overlap between the sequences. And in the second clade, it is shown that almost a third of genetic samples are considered to have the same genetic differences.
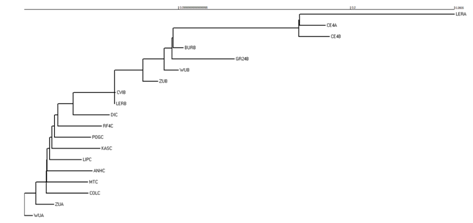
Figure 1 Phylogeny of Rpp8 Sequences. This is a phylogeny that was created from the Rpp8 sequences from the MEGA software. It was created using neighbor-joining techniques.
BEGIN TREES; |
ree Rpp8_exon_Inferred_Tree = ((((((((((((((((((((LERA:0.09000136624542679,CE4A:0.01503756983610329):0.000316165340808537,CE4B:0.01761276106787022):0.07334003045518941,BURB:0.00593938209990952):0.0005317053089915296,GR24B:0.03620311007347032):0.004668614044940492,WUB:0.008326044437385582):0.01239875306602262,ZUB:0.008793496401449212):0.01641548843393813,CVIB:0.0003676698446704323):0,LERB:0):0.02416450711484472,DIC:0.02111022109495939):0.008755805479867542,RF4C:0.02543199977040228):0.002005481802426233,POGC:0.02104453750789026):0.001505794741206088,KASC:0.02789363176748686):0.001389507151479656,LIPC:0.01857230438809743):0.001511738897326903,ANHC:0.02629285109028 ):5.742718077386632e-005,MTC:0.02371879148005516):0.0003265496464542152,COLC:0.02426326077083362):0.005977313377074652,ZUA:0.01037715260436323):0.006719285060534071,WUA:0.004599264671472049):0.004821488309967518,GR24A:0.02194269960138536):0.005586829289623845,BURA:0.01648827355939167,CVIA:0.01759480053849204); |
END; |
Table 1 Nexus File for Rpp8 Phylogeny
This is a phylogeny in nexus form that was created from the Rpp8 sequences from the MEGA software. It was created using neighbor-joining techniques.
The next step of the analysis was using the DNASP software. Something to note is that the final three bases in the gene sequences is the “stop” codon. This is generally not considered part of the coding region. As such, this paper excludes the last three bases for the purpose of analysis. The codon position of the first site is “1”. Five datasets were created: one consisting of the six A. thaliana A locus alleles, the second consisting of the six A. thaliana B (L.) Heynh locus alleles, the third consisting of the eight A. thaliana C locus alleles, the fourth combining the 20 A+B+C alleles, and the fifth consisting of the two A. lyrata alleles (Ce4A and Ce4B). Polymorphism analysis was conducted of the entire dataset, with the exclusion of the lyrata sequences. From the Polymorphic site test, there were 366 polymorphic sites identified out of 2664 total sites (Table 2). The DNA polymorphism test was also applied on the same set of data with the same restrictions. Pi was calculated to be 0.04005, k was 107, and theta was 103.164.
Polymorphic sites |
Input Data File: C:\...\Rpp8_exon.nex |
Population used: A_and_B_and_C |
Number of sequences used: 20 |
Selected region: 1-2748 Number of sites: 2748 |
Number of sites (excluding fixed gaps / missing data): 2742 |
Total number of sites (excluding sites with gaps / missing data): 2664 |
Sites with alignment gaps or missing data: 84 |
Invariable (monomorphic) sites: 2298 |
Variable (polymorphic) sites: 366 (Total number of mutations: 428) |
Singleton variable sites: 117 |
Parsimony informative sites: 249 |
Singleton variable sites (two variants): 109 |
Site positions: 22 37 180 302 305 350 351 353 357 387 564 695 765 |
816 868 941 970 1038 1058 1087 1111 1223 1297 1309 1317 1323 1346 |
1352 1398 1422 1435 1454 1468 1478 1479 1481 1482 1483 1485 1486 1487 |
1488 1500 1568 1569 1591 1621 1623 1630 1637 1638 1650 1661 1680 1697 |
1699 1753 1755 1759 1768 1791 1834 1853 1869 1882 1889 1902 1914 1916 |
1921 1923 1925 1928 1933 1944 1975 1980 2000 2017 2038 2041 2053 2054 |
2069 2078 2082 2113 2136 2188 2189 2195 2200 2237 2286 2294 2342 2371 |
2436 2440 2442 2489 2491 2510 2517 2558 2561 2575 2605 2743 |
Parsimony informative sites (two variants): 200 |
Site positions: 4 9 11 13 27 28 31 67 79 86 91 130 132 |
177 259 263 277 295 304 309 312 327 381 393 422 430 526 |
527 538 540 559 561 562 580 636 651 681 738 744 745 747 |
750 767 769 772 774 778 779 792 798 801 802 809 813 817 |
856 857 912 928 957 966 974 975 1112 1126 1166 1201 1203 1231 |
1282 1284 1299 1308 1312 1318 1327 1330 1332 1333 1342 1353 1384 1399 |
1416 1450 1457 1458 1459 1461 1466 1469 1470 1477 1491 1524 1553 1565 |
1594 1596 1639 1652 1659 1660 1684 1698 1706 1708 1766 1799 1840 1842 |
1849 1903 1906 1910 1924 1930 1931 1938 1963 1966 1978 1981 1982 1984 |
1994 1995 1997 1999 2008 2045 2052 2072 2093 2098 2108 2110 2112 2116 |
2117 2123 2138 2146 2168 2169 2187 2197 2204 2272 2273 2274 2277 2278 |
2284 2285 2291 2314 2325 2338 2339 2340 2343 2344 2347 2350 2355 2362 |
2370 2402 2409 2421 2424 2425 2441 2445 2448 2467 2468 2469 2472 2476 |
2490 2494 2495 2496 2503 2506 2509 2511 2514 2523 2573 2581 2610 2634 |
2637 2645 2648 2704 2744 |
Singleton variable sites (three variants): 8 |
Site positions: 255 343 1345 1898 1912 1922 2353 2357 |
Parsimony informative sites (three variants): 44 |
Site positions: 21 369 900 1143 1191 1291 1292 1303 1304 1316 1324 1325 1326 1328 1335 1355 |
1460 1462 1464 1467 1542 1685 1836 1843 1844 1850 1909 |
1915 1929 2051 2111 2194 2348 2354 2356 2419 2420 2426 2429 2431 2433 |
2572 2601 2644 |
Singleton variable sites (four variants): 0 |
Parsimony informative sites (four variants): 5 |
Site positions: 1334 1841 1845 1993 2428 |
Protein Coding Region assignation: No |
Table 2 Polymorphism Site Test
This table shows the results for the Polymorphism site test performed on the Rpp8 sequence data. There were 366 polymorphic sites and 2664 total sites used.
DNA polymorphism |
Input Data File: C:\...\Rpp8_exon.nex |
Population used: A_and_B_and_C |
Number of sequences used: 20 |
Selected region: 1-2748 Number of sites: 2748 |
Total number of sites (excluding sites with gaps / missing data): 2664 |
Number of polymorphic (segregating) sites, S: 366 |
Total number of mutations, Eta: 428 |
Number of Haplotypes, h: 20 |
Haplotype (gene) diversity, Hd: 1.000 |
Variance of Haplotype diversity: 0.00025 |
Standard Deviation of Haplotype diversity: 0.016 |
Nucleotide diversity, Pi: 0.04005 |
Sampling variance of Pi: 0.0000024 |
Standard deviation of Pi: 0.00156 |
Nucleotide diversity (Jukes and Cantor), Pi(JC): 0.04121 |
Theta (per site) from Eta: 0.04529 |
Theta (per site) from S, Theta-W: 0.03873 |
Variance of theta (no recombination): 0.0001722 |
Standard deviation of theta (no recombination): 0.01312 |
Variance of theta (free recombination): 0.0000041 |
Standard deviation of theta (free recombination): 0.00202 |
Finite sites model |
Theta (per site) from Pi: 0.04231 |
Theta (per site) from S: 0.04224 |
Theta (per site) from Eta: 0.04769 |
Average number of nucleotide differences, k: 106.689 |
Stochastic variance of k (no recombination), Vst(k): 2056.411 |
Sampling variance of k (no recombination), Vs(k): 232.418 |
Total variance of k (no recombination), V(k): 2288.829 |
Stochastic variance of k (free recombination), Vst(k): 35.563 |
Sampling variance of k (free recombination), Vs(k): 3.743 |
Total variance of k (free recombination), V(k): 39.307 |
Theta (per sequence) from S, Theta-W: 103.164 |
Variance of theta (no recombination): 1221.932 |
Variance of theta (free recombination): 29.07 |
Table 3 DNA Polymorphism Tests
This table displays the results from the DNA polymorphism test. Pi was calculated to be 0.04005, k was 107, and theta was 103.164.
After the basic divergence test, the DNASP software was then programmed to focus on the individual loci and the ways compare the individual loci. Looking at the A and B divergence, there were 12 sequences with 294 sequence sites and 325 total mutations. The k–value was ~105. The pi value was 0.035 (Table 4). The Dx/a value were 0.04 and the Da value is 0.02. Looking at the A and C divergence, there were 14 sequences with 302 polymorphic sites and 352 total mutations. The k–value was ~109, the pi value was ~0.036, the Dx/a = 0.03665, and the Da value is 0.00138 (Table 4). For the third comparison, the B loci were compared to the C loci. There were 14 total sequences with 346 polymorphic sites and 398 total mutations. The k–value was 110.648 and pi was equal to 0.04153 (Table 4). The Dx/y ratio was 0.04576 and the Da ratio was 0.01023 (Table 4). The final step of this analysis was using the DNASP using the HKA calculation method. By comparing the polymorphism within each species and the divergence observed between two species at two or more loci, the test can determine whether the observed difference is likely due to neutral evolution or rather due to adaptive evolution. To use the HKA calculator, we needed to determine how many segregating sites there were in the samples of each of the Rpp8 loci and the sample of the FAH1 alleles. It is already known how many segregating sites there were for the Rpp8 loci, so the attention was focused on the FAH1 alleles. The FAH1 alleles were found to have 10 segregating sites from DNA polymorphism testing (Table 5). After this, the amount of data was complete. For the HKA test, we had to calculate the number of differences between one A. lyrata allele and one A. thaliana allele for each of the loci (Rpp8 and FAH1). This was overcome by selecting a random A. lyrata gene and then finding its difference from each of the loci (Table 6). For the first locus, there were 159 differences. For the second locus, there were 334 differences (Table 6). This data was then put into the HKA test, which completed the requirements for the calculator. From the HKA–calculator, the p–value was calculated to be 0.0000, and the X–squared values was found to be 81.945 (Figure 2). Both of the calculations were done relative to the auto some (Table 7).
A vs B divergence |
Input Data File: C:\...\Rpp8_exon.nex |
Selected region: 1-2748 Number of sites: 2748 |
Total sites (excluding alignment gaps): 2676 |
Population 1: Six_A |
Number of sequences: 6 |
Number of polymorphic sites: 170 |
Total number of mutations: 183 |
Average number of nucleotide differences, k: 80.667 |
Nucleotide diversity, Pi(1): 0.03014 |
Population 2: Six_B |
Number of sequences: 6 |
Number of polymorphic sites: 181 |
Total number of mutations: 191 |
Average number of nucleotide differences, k: 85.800 |
Nucleotide diversity, Pi(2): 0.03206 |
Total data: |
Number of sequences: 12 |
Number of polymorphic sites: 294 |
Total number of mutations: 325 |
Average number of nucleotide differences, k: 105.803 |
Nucleotide diversity, Pi(t): 0.03954 |
Between populations: |
Number of fixed differences: 11 |
Mutations polymorphic in population 1, but monomorphic in population 2: 123 |
Mutations polymorphic in population 2, but monomorphic in population 1: 131 |
Shared Mutations: 60 |
Average number of nucleotide differences between populations: 124.611 |
Average number of nuc. subs. per site between populations, Dxy: 0.04657 |
Number of net nuc. subs. per site between populations, Da: 0.01546 |
Input Data File: C:\...\Rpp8_exon.nex |
Selected region: 1-2748 Number of sites: 2748 |
Total sites (excluding alignment gaps): 2676 |
Intraspecific Data: Six_A |
Number of sequences: 6 |
Interspecific Data: Six_B |
Number of sequences: 6 |
Number of segregating sites (Intraspecific Data), S: 170 |
Analysis of all sites |
Total number of sites analyzed: 2676 |
Polymorphism |
Nucleotide Diversity, Pi (Total): 0.03014 Pi (JC-Total): 0.03077 |
Theta (Total): 0.02995 Total number of substitutions: 183.00 |
Divergence |
Nucleotide Divergence, K (Total): 0.04657 K (JC-Total): 0.04807 |
A vs C Divergence |
Input Data File: C:\...\Rpp8_exon.nex |
Selected region: 1-2748 Number of sites: 2748 |
Total sites (excluding alignment gaps): 2703 |
Intraspecific Data: Six_A |
Number of sequences: 6 |
Interspecific Data: Eight_C |
Number of sequences: 8 |
Number of segregating sites (Intraspecific Data), S: 172 |
======== Analysis of all sites ======== |
Total number of sites analyzed: 2703 |
Polymorphism |
Nucleotide Diversity, Pi (Total): 0.03009 Pi (JC-Total): 0.03071 |
Theta (Total): 0.02997 Total number of substitutions: 185.00 |
Divergence |
Nucleotide Divergence, K (Total): 0.03665 K (JC-Total): 0.03757 |
Input Data File: C:\...\Rpp8_exon.nex |
Selected region: 1-2748 Number of sites: 2748 |
Total sites (excluding alignment gaps): 2703 |
Population 1: Six_A |
Number of sequences: 6 |
Number of polymorphic sites: 172 |
Total number of mutations: 185 |
Average number of nucleotide differences, k: 81.333 |
Nucleotide diversity, Pi(1): 0.03009 |
Population 2: Eight_C |
Number of sequences: 8 |
Number of polymorphic sites: 279 |
Total number of mutations: 315 |
Average number of nucleotide differences, k: 109.357 |
Nucleotide diversity, Pi(2): 0.04046 |
Total data: |
Number of sequences: 14 |
Number of polymorphic sites: 302 |
Total number of mutations: 352 |
Average number of nucleotide differences, k: 99.308 |
Nucleotide diversity, Pi(t): 0.03674 |
Between populations: |
Number of fixed differences: 0 |
Mutations polymorphic in population 1, but monomorphic in population 2: 37 |
Mutations polymorphic in population 2, but monomorphic in population 1: 167 |
Shared Mutations: 148 |
Average number of nucleotide differences between populations: 99.063 |
Average number of nuc. subs. per site between populations, Dxy: 0.03665 |
Number of net nuc. subs. per site between populations, Da: 0.00138 |
B vs C Divergence |
Input Data File: C:\...\Rpp8_exon.nex |
Selected region: 1-2748 Number of sites: 2748 |
Total sites (excluding alignment gaps): 2664 |
Intraspecific Data: Eight_C |
Number of sequences: 8 |
Interspecific Data: Six_B |
Number of sequences: 6 |
Number of segregating sites (Intraspecific Data), S: 270 |
======== Analysis of all sites ======== |
Total number of sites analyzed: 2664 |
Polymorphism |
Nucleotide Diversity, Pi (Total): 0.03980 Pi (JC-Total): 0.04090 |
Theta (Total): 0.04416 Total number of substitutions: 305.00 |
Divergence |
Nucleotide Divergence, K (Total): 0.04576 K (JC-Total): 0.04721 |
Input Data File: C:\...\Rpp8_exon.nex |
Selected region: 1-2748 Number of sites: 2748 |
Total sites (excluding alignment gaps): 2664 |
Population 1: Eight_C |
Number of sequences: 8 |
Number of polymorphic sites: 270 |
Total number of mutations: 305 |
Average number of nucleotide differences, k: 106.036 |
Nucleotide diversity, Pi(1): 0.03980 |
Population 2: Six_B |
Number of sequences: 6 |
Number of polymorphic sites: 177 |
Total number of mutations: 186 |
Average number of nucleotide differences, k: 83.267 |
Nucleotide diversity, Pi(2): 0.03126 |
Total data: |
Number of sequences: 14 |
Number of polymorphic sites: 346 |
Total number of mutations: 398 |
Average number of nucleotide differences, k: 110.648 |
Nucleotide diversity, Pi(t): 0.04153 |
Between populations: |
Number of fixed differences: 5 |
Mutations polymorphic in population 1, but monomorphic in population 2: 207 |
Mutations polymorphic in population 2, but monomorphic in population 1: 88 |
Shared Mutations: 98 |
Average number of nucleotide differences between populations: 121.896 |
Average number of nuc. subs. per site between populations, Dxy: 0.04576 |
Number of net nuc. subs. per site between populations, Da: 0.01023 |
Table 4 DNA Comparisons
This table lists the results from the divergence comparisons that were made on the Rpp8 gene sequences. In the table, there were two sets of tests performed on the data. The first one was a nucleotide-focused divergence test. And the second test is a DNA divergence and convergence statistical analysis. All of the possible comparisons among the three gene sequences were connected.
FAH1 |
Input Data File: C:\...\sequence_1.fas |
Selected region: 1-1563 Number of sites: 1563 |
Total sites (excluding alignment gaps): 912 |
Intraspecific Data: fhf |
Number of sequences: 57 |
Number of polymorphic (segregating) sites, S: 10 |
Genetic Code: Nuclear Universal |
Protein Coding, and Noncoding Regions analyzed: |
Number of protein coding regions (exons): 0 |
Number of noncoding regions (intronic and flanking regions): 1 |
Non coding region, from site: 1 to 1563 |
======== Analysis of silent sites ======== |
Total number of codons: 0 |
Number of codons analyzed: 0 ( 0 sites ) |
Total number of silent sites (synonymous and noncoding positions): 912.000 |
Total number of synonymous sites analyzed: 0.000 |
Total number of noncoding positions analyzed: 912 |
Polymorphism |
Nucleotide Diversity, Pi (Silent): 0.00351 Pi (JC-Silent): 0.00352 |
Theta (Silent): 0.00238 Number of silent substitutions: 10.00 |
Table 5 FAH1 DNA Polymorphism Test
This was a DNA polymorphism test that was done on the FAH1 alleles. The purpose of this test was to find the number of segregating sites in order to complete the HKA calculator. This test only looked at the 57 A. thaliana alleles. It did not include the Aly FAH1 allele. We see that the number of segregating sites expressed is 10.
Aly vs Loci one |
Input Data File: C:\...\Downloads\combined.fas |
Selected region: 1-2748 Number of sites: 2748 |
Total sites (excluding alignment gaps): 2712 |
Population 1: Alyrata |
Number of sequences: 2 |
Number of polymorphic sites: 70 |
Total number of mutations: 70 |
Average number of nucleotide differences, k: 70.000 |
Nucleotide diversity, Pi(1): 0.02581 |
Population 2: allele_one |
Number of sequences: 6 |
Number of polymorphic sites: 175 |
Total number of mutations: 188 |
Average number of nucleotide differences, k: 82.733 |
Nucleotide diversity, Pi(2): 0.03051 |
Total data: |
Number of sequences: 8 |
Number of polymorphic sites: 384 |
Total number of mutations: 419 |
Average number of nucleotide differences, k: 159.036 |
Nucleotide diversity, Pi(t): 0.05864 |
Between populations: |
Number of fixed differences: 165 |
Mutations polymorphic in population 1, but monomorphic in population 2: 66 |
Mutations polymorphic in population 2, but monomorphic in population 1: 184 |
Shared Mutations: 4 |
Average number of nucleotide differences between populations: 261.833 |
Average number of nuc. subs. per site between populations, Dxy: 0.09655 |
Number of net nuc. subs. per site between populations, Da: 0.06839 |
Aly vs Loci two |
Input Data File: C:\...\Downloads\combined.fas |
Selected region: 1-2748 Number of sites: 2748 |
Total sites (excluding alignment gaps): 903 |
Population 1: Alyrata |
Number of sequences: 2 |
Number of polymorphic sites: 24 |
Total number of mutations: 24 |
Average number of nucleotide differences, k: 24.000 |
Nucleotide diversity, Pi(1): 0.02658 |
Population 2: alleletwo |
Number of sequences: 57 |
Number of polymorphic sites: 671 |
Total number of mutations: 680 |
Average number of nucleotide differences, k: 310.633 |
Nucleotide diversity, Pi(2): 0.34400 |
Total data: |
Number of sequences: 59 |
Number of polymorphic sites: 839 |
Total number of mutations: 1179 |
Average number of nucleotide differences, k: 333.991 |
Nucleotide diversity, Pi(t): 0.36987 |
Between populations: |
Number of fixed differences: 478 |
Mutations polymorphic in population 1, but monomorphic in population 2: 21 |
Mutations polymorphic in population 2, but monomorphic in population 1: 677 |
Shared Mutations: 3 |
Average number of nucleotide differences between populations: 663.719 |
Average number of nuc. subs. per site between populations, Dxy: 0.73502 |
Number of net nuc. subs. per site between populations, Da: 0.54973 |
Table 6 DNA Polymorphism Difference between Random Allele and A. lyrata
This table shows the results from the polymorphism difference test. This was conducted in order to find the difference between these two allele types for the HKA calculator.
HKA test. Direct mode |
Locus 1 |
Locus 1 |
Interspecific polymorphism data |
||
Segregating sites (obs) |
366 |
10 |
Segregating sites (exp) |
234.05 |
141.95 |
Total number of sites |
2664 |
912 |
Sample size |
2664 |
912 |
Interspecific Divergence |
||
No. differences (obs) |
159 |
334 |
No. differences (exp) |
290.95 |
202.05 |
Total number of sites |
2664 |
912 |
Chromosomal location |
Autosome |
Autosome |
X-square value |
81.945 |
P: 0.0000 |
Table 7 Results from the Direct HKA Calculator
This table shows the results of the direct HKA calculator. The X-square value is ~82, ad the P-value is ~0.
From the neighbor–joined phylogeny, there is a significant amount of evidence for interlocus exchange. In the phylogeny, every branch shares significant similarities with other branches within and in different clades. Also, the phylogeny is in two clades. There is one “outlier,” WUA, that shares remarkable similarities with the rest of the gene sequences. And the second clade is one big clade that devolves into smaller clades. This de–evolution of the clade design shows significant amount of genetic overlap between the sequences. And in the second clade, it is shown that almost a third of genetic samples are considered to have the same genetic differences. All of these are traits of interlocus exchange. Because of the exchange, the sequences share a significant amount of similarities. Overall, the phylogeny tree suggests sharing or exchange of alleles/polymorphism between the A, B and C loci. These similarities influence the shape of the phylogeny, making all of the branches and clades relatively similar.
Around ~5% of the segregated DNA polymorphism is shared between loci and 95% is distinct to each locus. This can be done by directly comparing the number of shared and locus–specific segregating sites and/or by comparing the within–and between–locus nucleotide diversity. This paper finds this amount by finding the average of pi between all three of the loci comparisons (Table 5). Pi from A vs B is 0.03954. Pi from A vs C is 0.0367. And Pi from B vs C is 0.04153. Taking the average and multiplying by 100, we get a similarity rate of about 5%. And we can find the distinction rate by doing 1– 5%, which gives us 95%. There is evidence for interlocus exchange. This is shown by looking at the pi values and the k–values for the nucleotide diversity. All of the pi values for each of the comparisons is between 3–6% (Table 5). This is significant as this means that 3–6% of the nucleotides are different when being compared to a chromosomal paralog. As such, this supports the idea that these genes exist within a interlocus exchange. This is because the genes are still mainly similar, while also sharing significant differences. And because of the similarity between all three of these genes, the differences coming from one gene might be coming from both of its chromosomal paralogs. This explains why the pi value for all of the genes is somewhat constant, as it characterized by an interlocus exchange. Intergenic exchange is more common between B and C loci than it is between any other loci combination. This was obtained by looking at the pi and k values of all of the possible combinations of loci. Pi from A vs B is 0.03954. Pi from A vs C is 0.0367. And Pi from B vs C is 0.04153 (Table 4;5) Because of this, B vs C pi’s is the greatest number. This is significant because that means that B vs C have the greatest amount of genetic differences. And since there is evidence that these differences are mainly coming from interlocus exchanges, then this supports the idea that intergenic exchange between the B and C loci is more common than any other combination. Otherwise, this particular combination wouldn’t have the largest pi value.
However, we must keep in mind that Arabidopsis is normally self–fertilizing, so almost all individuals in nature will be homozygous for an identical chromosome. In order to account for this context, we must hypothesize how theoretically ringing intergenic recombination propensities would correlate without date. In line with the data, intergenic recombination propensities would be dictated by the homozygous nature of the chromosomes. This would allow for less diversity in gene sequences and less influential recombination. The recombination propensity would also focus on the B and C loci. This is because they are shown to be the center of genetic changes out of the three possible recombination events. As such, the intergenic recombination would share propensities with these two loci. These propensities might include the direction of the recombination and the polarity of the dividing cell. All of these characteristics are influential in the process and serve to change the final recombination. There is elevated polymorphism at Rpp8. From the HKA–calculator the p–value was calculated to be 0.0000, and the X–squared values was found to be 81.945 (Table 6). Both of the calculation was done relative to the auto some (Table 6). Because the p–value is 0, this means that the association between the increased polymorphism and the presence of Rpp8 is statistically significant. From the gene sequences discussed here, it seems that these significant values came from too much polymorphism among the loci at nonsynonymous sites. This provides evidence of a positive directional flow, meaning that there is elevated polymorphism at Rpp8. All of the increased of the HKA–calculator to the p–value indicates that there is an elevated polymorphism at Rpp8. This elevated polymorphism has some potential causes. A polymorphism can be maintained by a balance between variation created by new mutations and natural selection. Genetic variation may be caused by frequency–dependent selection. Multiple niche polymorphisms exist when different genotypes should have different fitnesses in different niches. Heterozygous advantage may maintain alleles which would other be selected against. If selection is operating, migration can introduce polymorphism into a population. For this specific population, it is likely that the polymorphism demonstrated in this paper is coming from a continuously maintained balance of variation created by new mutations and natural selection. This seems to fit with the context of the gene sequences, as they are coming from Arabidopsis, which is self–fertilizing. This means that almost all individuals in the sample will be homozygous for an identical chromosome. This allows for there to exist such a delicate balance, thus impacting the polymorphism.
From the neighbor–joined phylogeny, there is a significant amount of evidence for interlocus exchange. Around ~5% of the segregated DNA polymorphism is shared between loci and 95% is distinct to each locus. There is evidence for interlocus exchange. Intergenic exchange is more common between B and C loci than it is between any other loci combination. There is elevated polymorphism at Rpp8. All of these observations are coming from the hypothesis that the polymorphism demonstrated in this paper is coming from a continuously maintained balance of variation created by new mutations and natural selection. This seems to fit with the context of the gene sequences, as they are coming from Arabidopsis, which is self–fertilizing. Some future directions of research include looking for the cause of the increased polymorphism in Rpp8. Even though this paper shows the link, it is still unknown why this relationship exists with this specific gene.16 More work should be done to clarify this relationship and compare it to similar relationships with other genes. Also more work should be done to find molecular evidence for this relationship. An assay should be done in order to account for the entire normal gene irregularities, which would help give support to the hypothesis discussed here today. Also, more work should be done on other species. Doing more work on other species will assist the relationship presented in this paper. And more work should be done on the phylogenetics of Rpp8.17 It would be beneficial to see a detailed look and phylogeny of how the gene has evolved throughout different contexts and how this is reflected in the data. And a final suggestion would be to clarify the intergenic recombination propensities. These were conjectured within this paper based off of data, but more molecular data needs to be shown for this hypothesis.
None.
The author declares there is no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.