Journal of
eISSN: 2377-4282


Horseradish peroxidase nanoparticles was immobilized covalently onto inner wall of polyvinyl chloride (PVC/plastic) vial through glutaraldehyde coupling with 64.13% retention of its initial activity and conjugation yield of 0.35mg/cm2. The vial bound enzyme nanoparticles showed optimum activity at pH-7.2, when incubated at 40ºC for 5min. There was a linear relationship between immobilized peroxidase nanoparticles activity and H2O2 concentration in the range of 0.01mM to 100mM with apparent Km for H2O2 as 20mM. The vial bound enzyme nanoparticles was employed for enzymic colorimetric determination of H2O2 in milk and urine. Minimum detection limit of 0.01mM was obtained. Analytical recoveries of added H2O2 in urine (10mM and 20mM) were observed by 98.0% and 96.5% respectively. Within and between assays coefficients of variation (CVs) for H2O2 in urine determinations were detected by 5.3% and 4.5% respectively. A good correlation (r=0.997) was obtained between H2O2 values by standard enzymatic colorimetric method using free enzyme and the present method. The polyvinyl chloride vial was reused 150 times over a period of 4 months, when stored at 4oC. The method has the advantage that it provides the reusability of immobilized enzymes nanoparticles with enormous ease.
Keywords: Horseradish peroxidase, Nanoparticles, Plasticized polyvinyl chloride vial, Immobilization, Milk, Urine
PVC, Polyvinyl Chloride; SEM, Scanning Electron Microscopy; CVs, Coefficients of Variation
Peroxidases are a large family of heme-containing enzymes that catalyze oxidation and reduction reactions of enormous families of substrates. These substrates are specific and have potential applications in therapeutics and clinical uses.1 Peroxidases are capable of oxidizing the functional group R-N=N-R′ (azo-dyes) in which R and R′′ can be either aryl or alkyl groups. Hence, peroxidases work as antioxidents and extensively used in clinical and immunological analysis including glucose, cholesterol, and urea determinations.2 Horseradish peroxidase is one of the most valuable marker for oxidative stress and is associated with many pathological conditions causing diabetes, atherosclerosis and aging. Horseradish peroxidase is used in variety of organic compounds as donors and acceptors of electron.3 Different approaches were previously reported for detection and measurement of hydrogen peroxide including photometric, idiometric, colorimetric, spectrophotometric and chemiluminescence methods.4 Immobilization of the enzymes on an insoluble support had many advantages over the use of free enzymes such as enzyme reusability, improved stability, continuous operation, controlled product formation, simplified and efficient processing and often economy via glutaraldehyde coupling.5 or diazotization.6 Modification of polymers on surface was performed by the introduction of active functional groups for covalent attachment of biomolecules such as protein, DNA and carbohydrates.7 Plasticized polyvinyl chloride (PVC) has exceptional chemical and physical properties providing an ideal support for direct immobilization of enzyme after its chemical cleavage.8 Considering these advantages, various enzymes were immobilized onto PVC sheet. The immobilization of Candida Cylindracea lipase was therefore done on Polyvinyl chloride chitin and agarose.9 Covalent immobilization of enzymes onto surface of a polyvinyl chloride (PVC) sheet is one of the widely available, economic, chemically inert, corrosion free, tough, light weight and maintenance free. PVC provides a high strength-to-weight ratio which does not allow leakage of enzyme.10 In addition, Horseradish peroxidase was co-immobilized with lipase, glycerol kinase and glycerol-3-phosphate oxidase. A covalent coupling method was applied for plasticized polyvinyl chloride (PVC) strip immobilization and was further used for enzyme colorimetric determination of serum triglyceride.11 In another method, HRP was co-immobilized with lipase, glycerol kinase, glycerol-3-phosphate oxidase onto aryl amine glass beads affixed on plastic strip and used for determination of triglycerides in serum.12 Furthermore, the immobilization of horseradish peroxidase onto chemically activated chitin was reported.13 In our present work, immobilization of horseradish peroxidase nanoparticles was performed onto cost effective and easy-to-prepare plasticized activated polyvinyl chloride vial and studied for estimation of H2O2 in milk and urine samples.
Activation of plasticized polyvinyl chloride vial
The plasticized polyvinyl chloride vial was first incubated with nitrating acid (mixture of conc. HNO3 and H2SO4 in 5:1 ratio) for 24 hours to cleave oxidative polyvinyl chloride polymers into small chain polymers having protruding ends toward the surface. Then, this acid-treated plasticized polyvinyl chloride vial was rinsed with distilled water repeatedly after which the vial was incubated with 2.5% (w/v) glutaraldehyde solution in 0.05 M sodium phosphate buffer (pH 7.0) for 48 hours at room temperature.5,7,9
Preparation of horseradish peroxidase nanoparticles
The horseradish peroxidase nanoparticles were synthesized by desolvation method. To horseradish peroxidase enzyme solution (1 mg ml−1), 5 ml of absolute ethanol was added drop wise at a rate of 0.1-0.2 ml min−1 under continuous stirring at a speed of 8,000×g. The desolvating agent encouraged the enzyme/protein molecules to aggregate into small particles by removing in between water molecules, thus reducing the distance between the enzyme molecules. This was followed by an addition of 1 ml glutaraldehyde solution (1 %v/v) in the above solution under the similar stirring condition at 4°C for 24 h to ensure complete cross-linking of horseradish peroxidase nanoparticles. Such a high concentration of glutaraldehyde is likely to provide intermolecular cross-linking of horseradish peroxidase molecules through Schiff base. The enzyme nanoparticles thus formed were separated from free enzyme by centrifuging enzyme nanoparticles suspension at 15,000×g for 10 min at 4°C, followed by redispersion of horseradish peroxidase nanoparticles in 50mM of phosphate buffer (pH 7.4) and sonication for 5 min.
Immobilization of horseradish peroxidase enzyme nanoparticles onto activated plasticized polyvinyl chloride vial
The activated polyvinyl chloride vial was incubated with 3ml of horseradish peroxidase nanoparticles at 4ºC for 48h in dark. The excess enzyme nanoparticles was decanted off and tested for activity and protein concentration. The horseradish peroxidase nanoparticles bound activated plasticized polyvinyl chloride vial was later stored at 4ºC.7,9
Assay of activated plasticized polyvinyl chloride vial bound horseradish peroxidase nanoparticles
The enzyme assay based on trinder’s color reaction was carried out by incubating it for 10 minutes at 25ºC with the mixture of 2.8ml sodium phosphate buffer (0.05M) pH 7.0, 0.1 ml 4-aminophenazone and 0.1 ml phenol with addition of 0.1ml of 10mM peroxide solution in dark and read at A520 was read against control. Its conjugation yield and % retention of enzyme nanoparticles activity after immobilization was measured.7,9
Kinetic characterization
The optimal pH, time of incubation and temperature for immobilized horseradish peroxidase nanoparticles were determined for its maximal activity by varying pH of 0.2M sodium phosphate buffer (pH-6.8,7.0,7.2,7.4, 7.6,7.8,8.0), time (5minutes-30minutes) with an interval of 5 minutes and temperature range of 25C-45°C (with an interval of 5°C), respectively, and read at A520. The effect of substrate concentration on activity of immobilized enzyme nanoparticles was studied by performing the enzyme assay at different hydrogen peroxide concentration (0.01 mM-100 mM) by trinder’s color reaction and read at A520. The interfering effect of various metals and metabolites is found in blood such as iron, copper, zinc, potassium, sodium, urea, ascorbic acid, cholesterol and glucose were studied at their physiological concentrations to studied their effects on immobilized enzyme nanoparticles.7,9
Evaluation of activated plasticized Polyvinyl chloride vial bound horseradish peroxidase nanoparticles
The method was evaluated by studying its analytical recovery and precision. The urine H2O2 values obtained by present method (y) were compared with those by standard enzymatic colorimetric method employing horseradish peroxidase nanoparticles (x). Their correlation coefficient (r) was calculated using the regression equation.4,5,7,9
Applications to the determination of H2O2 in milk samples
The activated plasticized polyvinyl chloride vial bound horseradish peroxidase nanoparticles was used to detect the hydrogen peroxide concentration in milk samples. Samples of the sterilized dry milk, sterilized whole milk which were taken in homogenized form and raw milk were collected. The samples were added in activated plasticized polyvinyl chloride vial bound horseradish peroxidase nanoparticles and the enzyme assay was done by trinder’s color reaction.7,9
Reusability and storage stability of activated plasticized polyvinyl chloride vial bound horseradish peroxidase nanoparticles
The enhanced storage and stability of activated plasticized polyvinyl chloride vial bound enzyme nanoparticles was studied over a period of 4 months during its storage 4ºC. The immobilized horseradish peroxidase nanoparticles activity was measured by trinder’s color reaction at every week.7,9
%Immobilization of horseradish peroxidase nanoparticles onto activated plasticized polyvinyl chloride vial
Horseradish peroxidase nanoparticles was immobilized covalently onto inner wall of a activated plasticized polyvinyl chloride vial with a conjugation yield of 0.35mg/cm2 and 64.13% retention of its initial activity. During immobilization, vinyl polymers of plasticized polyvinyl chloride were spilled down by treating it with HNO3 and H2SO4 mixture which introduced nicks in long-chain polymer and generated free ends protruding from the polymer surface. These free ends were reacted with the aldehyde group of glutaraldehyde, one aldehyde group of a glutaraldehyde reacted with free end of vinyl chain to form a –C=CH- bond between plasticized PVC sheet and glutaraldehyde. That reaction process was lead to surface activation for covalent immobilization of enzyme nanoparticles. The -NH2 group on the surface of enzyme nanoparticles was attached covalently onto other –CHO group of glutaraldehyde, already attached to plasticized PVC sheet, through Schiff base formation. This led to covalent immobilization of peroxidase nanoparticles with plasticized PVC vial surface.8 The Scanning Electron Microscopy (SEM) of activated plasticized polyvinyl chloride vial surface with the immobilized enzyme nanoparticles was coined change in surface morphology which showed folds and clusters along with some beaded structures that were not observed in plasticized PVC sheet alone (Figure 1). A change in surface morphology of the support after immobilization confirmed enzyme immobilization.8
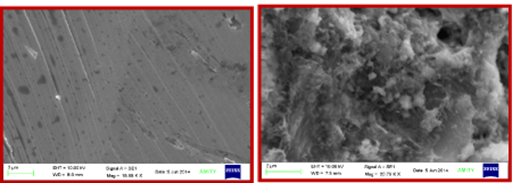
Figure 1 Scanning electron micrographs of activated plasticized polyvinyl chloride vial surface
a. without immobilized horseradish peroxidase nanoparticles
b. with immobilized horseradish peroxidase nanoparticles. Evaluation planning and implementation.
Studied kinetic characterization of activated plasticized polyvinyl chloride vial bound horseradish peroxidase nanoparticles
The horseradish peroxidase nanoparticles immobilized polyvinyl chloride vial was showed optimum activity at pH-7.2, which was higher as compared to the optimum pH of 7.0 for free enzyme.14 Li and Townshend.15 was also reported different optimum pH for the peroxidase activity before (7.0) and after immobilization (5.8) on polytetrafluoroethylene tubing. Comparable results were reported for peroxidase immobilized on sol-gel matrices.16 and porous glass.17 respectively. Substrate saturation was observed for activated plasticized polyvinyl chloride vial bound peroxidase at 100mM, which was demonstrated by Line-weaver Burk plot. Km of immobilized enzyme nanoparticles was found to be 20mM (Figure 2), indicating the increased affinity of immobilized enzyme for its substrate. Km values for immobilized peroxidase nanoparticles bound PVC in the present study are also lower than the reported Km values for peroxidase immobilization onto clay minerals.18 modified chitosan beads.19 and activated wool.20 indicating the suitability of plasticized polyvinyl chloride vial as enzyme nanoparticles immobilization support.
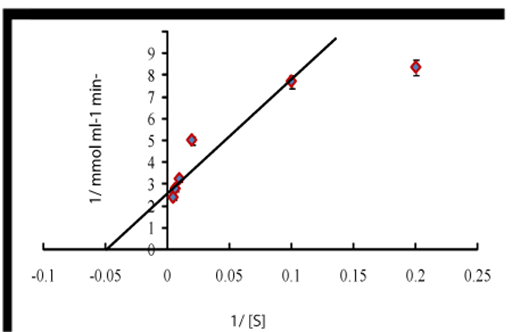
Figure 2 A Lineweaver–Burk plot between 1/[S] and 1/[v] of activated plastiziced polyvinyl chloride vial bound horseradish peroxidase nanoparticles.
The optimum temperature for immobilized peroxidase nanoparticles was 40°C, as compared to free enzyme which shows optimum activity at 25°C.14 This change signifies the protecting effect of support which might have stabilized the three dimensional structure of enzyme. An increase in the optimum temperature of peroxidase after immobilization has also been reported in literature such as on activated wool (40°C).20 magnetic beads (35°C).21 and modified chitosan beads (45°C).19
The change in the kinetic parameters of the immobilized enzymes nanoparticles might be due to change in the microenvironment or conformation of the enzymes nanoparticles after getting immobilized.4,5,7,9 Hence, this result was showed that there was practically no interference by these metabolites contrary to earlier report shows interference by ascorbic acid.22
Evaluation of activated plasticized polyvinyl chloride vial bound horseradish peroxidase nanoparticles for H2O2 determination
Effect of substrate concentration on the activity of immobilized horseradish peroxidase nanoparticles was studied in the concentration range from 0.01mM to 100mM. The effect of varying concentration of H2O2 on initial velocity of peroxidases was hyperbolic from 0.01mM to 100mM, after which the reaction rate remained constant. The minimum detection limit of the method was found to be 0.01mM in reaction mixture, which was better than those employing enzyme immobilized on to activated wool.20 and modified chitosan.19 To check the reliability of the method, the analytical recovery of added H2O2 in the six urine samples was determined. The mean analytical recoveries of added H2O2 (10mM and 20mM) in urine were 98.0% and 96.5% respectively. To study the reproducibility and reliability of the method, the H2O2 content in six urine samples in one run (within batch) and after storage at 4oC for one week (between batch) were determined. The results showed that the H2O2 value of this determination agreed with each other and within batch and between batch coefficients of variation (CVs) were 5.3% and 4.5%. The H2O2 level in 10 urine samples from apparently healthy adults and gout patients as determined by the present method (y) were compared with those by standard enzymatic colorimetric kit method (x). The values obtained by both the methods agreed with each other showing a good correlation (r=0.997) (regression equation being y=1.0148x+0.0072) (Figure 3).23
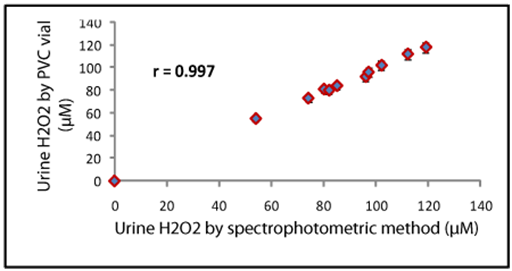
Figure 3 Correlation between urine H2O2 values as determined by the spectrophotometric method employing free horseradish peroxidase (x-axis) and the present method (y-axis) using activated plasticized polyvinyl chloride vial bound horseradish peroxidase nanoparticles.
Applications to the determination of H2O2 in milk samples
The developed method of the activated plasticized polyvinyl chloride vial bound horseradish peroxidase nanoparticles under its assay conditions was used in estimation of H2O2 level in selected milk samples. The procedure was same as described in the materials and methods section except that the H2O2 was replaced with milk samples and absorbance of testing solutions after enzyme assay was read at A520. The results are fairly good when compared with the reference method (Table 1).23
Types of milk samples |
Estimated H2O2 level (μM) (mean±SE) |
Sterilized dry milk |
85±0.04 |
Sterilized whole milk |
72±0.03 |
Raw milk |
48±0.06 |
Table 1 Estimated hydrogen peroxide level in selected milk samples by using activated plasticized polyvinyl chloride vial bound horseradish peroxidase nanoparticles based on trinder’s color reaction which was read at A520
Storage stability and reusability of activated plasticized polyvinyl chloride vial bound horseradish peroxidase nanoparticles
The activated plasticized polyvinyl chloride vial bound horseradish peroxidase nanoparticles lost 50% of its initial activity after its 150 regular use over a period of 4 months (Figure 4), when stored in sodium phosphate buffer (pH 7.2) at 4oC was found to be higher as compared to immobilized enzyme onto chitosan.12 aluminum-pillared inter layered clay.24 and Sephadex G-50.25 Free enzyme retained only 20% of the initial activity after one month.14 clearly suggesting the stabilizing effect of immobilization on the enzyme.
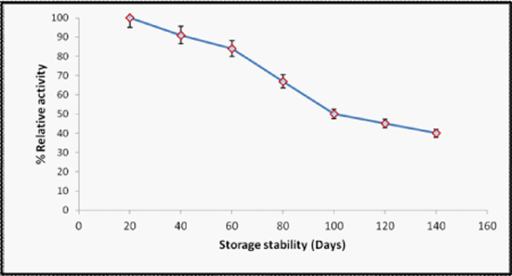
Figure 4 Effect of storage stability relative to enzyme activity of activated plasticized polyvinyl chloride vial bound horseradish peroxidase at 4ºC
The activated plasticized polyvinyl chloride vial bound horseradish peroxidase nanoparticles retained 64.13 % of its initial activity with a conjugation yield of 0.35 mg protein cmˉ2. Hence, this developed method has the advantage over free enzyme which provides maximal reuse of immobilized enzyme nanoparticles with enormous ease up to 150 times over a period of 4 months to reduce its cost over testing as well as with its enhanced storage stability at 4oC. So, overall interpretations were showed that our developed method of immobilization of horseradish peroxidase nanoparticles on to activated plasticized polyvinyl chloride vial via its chemical modification with glutaraldehyde coupling was cost effective method and easy to perform with reduced labor as well as minimal requirements of expensive chemicals and instrumentation too as compared to other conventional and advanced methods.
The present work was supported by SERB, Department of Science and Technology (DST), India. Thanks to all scientists referenced throughout the paper whose valuable work has guided the way throughout this research works
None.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.