Journal of
eISSN: 2373-4469


Review Article Volume 2 Issue 5
1Division of Neurogenetics, Nagoya University Graduate School of Medicine, Japan
2Cold Spring Harbor Laboratory, USA
Correspondence: Kinji Ohno, Division of Neurogenetics, Nagoya University Graduate School of Medicine, Japan, Tel +81527442446, Fax +81527442449
Received: October 21, 2015 | Published: November 27, 2015
Citation: Rahman MA, Ohno K. Splicing aberrations in congenital myasthenic syndromes. J Investig Genomics. 2015;2(6):111-118. DOI: 10.15406/jig.2015.02.00038
The fidelity of splicing is indispensable for assuring rapidly changing cellular processes and physiological integrity of the cells. The precise biogenesis of ribo nucleoprotein complexes (RNPs) is therefore essential, which is ensured by proper complementation of RNA and RNA-binding proteins in a specific cellular context. Genetic mutations disrupting cis-acting splicing elements or compromising catalytic functions of trans-acting RNA-binding proteins can impair the biogenesis of functional RNPs and provoke pathological consequences. Analyses of splicing aberrations in human diseases not only allow us to understand the underlying maladies in pathological conditions, but also pave the way to gain insight into splicing regulation under physiological conditions. Dissection of the patho mechanisms of splicing defects in neuromuscular junction (NMJ) disorders has disclosed unprecedented roles of RNA-binding proteins in the neuromuscular signal transmission at the NMJ. This review focuses on splicing defects in congenital myasthenia syndromes (CMS), hereditary disorders caused by germ line mutations in molecules expressed at the NMJ. Splicing mis-regulations of the NMJ molecules that have been uncovered in the course of analyses of splicing mutations causing CMS can be extrapolated to other diseases caused by germ line or somatic mutations.
Keywords: neuromuscular junction, congenital myasthenic syndromes, alternative splicing, splicing cis-elements; splicing trans-factors, spliceosome, mutation, aberrant splicing
NMJ, neuromuscular junction; CMS, congenital myasthenic syndromes; NMD, nonsense-mediated mrna decay; BP, branch point; PPT, polypyrimidine tract; ISE, intronic splicing enhancer; ESE, exonic splicing enhancer; ISS, intronic splicing silencer; ESS, exonic splicing silencer; SR Proteins, serine/arginine-rich proteins; hnRNPs, heterogeneous nuclear ribo nucleoproteins; AChE, acetyl cholinesterase; AChR, acetylcholine receptor; PTC, premature termination codon; NAS, nonsense-associated altered splicing; NASRE, nonsense-associated altered splicing of a remote exon; TE, transposable elements; Sines, short interspersed nuclear elements; MIRs, mammalian interspersed repeats; Fz-CRD, frizzled-like cysteine-rich domain; AON, antisense oligo nucleotide
Neuromuscular junction (NMJ) is the synapse between the motor neuron and the muscle fibers, which transmits signals from neuron to muscle to ensure proper muscle contractions. A number of molecules constitute the complex molecular architecture of the NMJ and a series of signaling cascades are involved in the formation of the NMJ (Figure 1). Congenital myasthenic syndromes (CMS) are a group of NMJ disorders, which compromise the safety margin of neuromuscular signal transmission. CMS arise due to defects in genes coding for presynaptic, synaptic, and postsynaptic molecules at the NMJ (Figure 1), and constitute a group of clinically heterogeneous disorders presenting with abnormal muscle fatigue, muscle weakness, amyotrophy and sometimes minor facial anomalies.1,2 CMS patients show variable patterns of affected muscles, variable ages of onset with dominance of infancy, and variable responses to pharmacological agents. CMS are mostly caused by autosomal recessive loss-of-function mutations. An autosomal dominant gain-of-function mutation is observed only in the slow-channel syndrome, in which openings of acetylcholine receptor (AChR) are abnormally prolonged. Some loss-of-function mutations in CMS are due to mutations affecting the fidelity of splicing. As observed in splicing mutations in other diseases, splicing errors in CMS are comprised of aberrant inclusion of a non-functional exon, aberrant skipping of an exon encoding the important functional domain, aberrant retention of an intron, alternative selection of a 5’ or 3’ splice site, activation of a cryptic splice site, or aberrant splicing due to a premature termination codon (PTC) in a remote exon. In this review, we address numerous modes of aberrant splicing in CMS, which affect the physiological integrity of the NMJ and compromise the safety margin of the neuromuscular signal transmission. In this review, we use the legacy annotation for AChR subunit genes, so that mutations in this review and the references can be readily traced. The AChR α (CHRNA1) and ɛ (CHRNE) subunits have a signal peptide comprised of 20 amino acids at the N-terminal end. As position 1 in the legacy annotation is the N-terminal end of a mature peptide, p.Glu174Ter in CHRNE in the canonical annotation, for example, is indicated as ɛE154X. Similarly c.613-619del7 in CHRNE is indicated as ɛ553del7. As the other genes do not encode a signal peptide, mutations in the other genes (COLQ, DPAGT1, and DOK7) are indicated according to the canonic annotation.
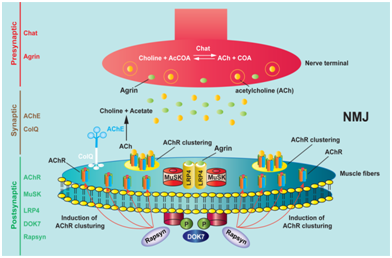
Figure 1 Schematic presentation of the molecular architecture of neuromuscular junction (NMJ) and neuromuscular signal transmission. Upon arrival of a motor nerve terminal near the muscle fibers, agrin is released from the nerve terminal. Agrin interacts with LRP4 (low-density lipoprotein receptor-related protein 4) and stimulates the association of LRP4 and MuSK (muscle-specific receptor tyrosine kinase) to induce MuSK phosphorylation. Activation of MuSK is enhanced by an adaptor protein, DOK-7. Activation of MuSK induces clustering of acetylcholine receptor (AChR) on the scaffold of self-aggregated rapsyn. AChR clusters are thus highly concentrated at the postsynaptic membrane. Presynaptic, synaptic and postsynaptic regions are marked on the left side, and essential molecules in each region are indicated.
Physiology of pre-mRNA splicing
Pre-mRNA splicing is carried out in the nucleus by a macromolecular complex called splicesome, which is composed of RNA and proteins. Recognition of intron/exon boundary is guided by essential splicing regulatory cis-elements close to either end of an intron, which are termed the consensus splice site sequences. These include the 5’ splice site (a splice donor site), the branch point (BP), the polypyrimidine tract (PPT), and the 3’ splice site (a splice acceptor site). The assembly of splicesome is further modulated either in a positive and negative manner by auxiliary splicing cis-elements termed intronic/exonic splicing enhancers (ISEs/ESEs) and intronic/exonic splicing silencers (ISSs/ESSs), respectively. Most of these auxiliary cis-elements function by binding to cognate proteins (trans-factors), whereas some cis-elements function by forming a secondary structure. The majority of splicing trans-factors for ESE are serine/arginine-rich (SR) proteins, whereas the majority of splicing trans-factors for splicing silencer elements (ISSs/ESSs) are heterogeneous nuclear ribonucleic proteins (hnRNPs).3 Among the four classes of cis-elements, ISEs are the least characterized. However, hnRNP F, hnRNP H, NOVA1, NOVA2, FOX1, and FOX2 have recently been identified as potential candidate trans-factors for ISEs.4‒7
Mutations affecting the consensus splicing cis-elements
About 15 to 20% of human genetic diseases are caused by mutations that disrupt the consensus or auxiliary splicing cis-elements.8 The majority of splicing mutations are at the consensus splicing cis-element, and decrease the binding affinity for an essential trans-factor, which gives rise to compromised recognition of a splice site. In CMS, we and others have reported mutations at the consensus splice sites causing aberrant splicing. Representative mutations are shown in Table 1. For example, ɛIVS6+1G>T at the 5’ splice site of CHRNE intron 6 causes skipping of exon 6 (Figure 2b).9 Another 5’ splice site mutation, ɛIVS7+2T>C in CHRNE intron 7, causes retention of intron 7.10 On the other hand, ɛIVS9-1G>C at the 3’ splice site of CHRNE intron 9 causes retention of intron 9, and subsequently generates a PTC in exon 10 in the final transcript.11 Another 3’ splice site mutation in CHRNE intron 4 is ɛIVS4-2A>C, which causes skipping of exon 5, thereby deletes codons 96 to 147 in the final transcript (Figure 2c).11
Mutations affecting the auxiliary splicing cis-elements (ISEs/ESEs/ISSs/ESSs)
Gene |
Mutation |
Molecular Consequence |
CHRNE9 |
ɛIVS6+1G>T |
Disrupts the 5’ splice site of intron 6, which causes skipping of exon 6 |
CHRNE 10 |
IVS7+2T>C |
Disrupts the 5’ splice site of intron 7, which causes retention of intron 7 |
CHRNE17 |
IVS6-1G>C |
Disrupts the 3’ splice site of intron 6, which activates a cryptic splice site |
CHRNE11 |
ɛIVS9-1G>C |
Disrupts the 3’ splice site of intron 9, which causes retention of intron 9. This generates a premature termination codon (PTC) in exon 10 |
CHRNE11 |
ɛIVS4-2A>C |
Disrupts the 3’ splice site of intron 4, which causes skipping of exon 5.This results deletion of codons 96-147 |
p.E415G |
Disrupts an ESE, and de novo generates an ESS. This causes skipping of exon 16 and a shift of downstream frame |
|
CHRNA1 22 |
P3A23’G>A |
Disrupts an ESS, and de novo generates an ESE. This causes inclusion of a non-functional exon P3A, which disables expression of acetylcholine receptors at the cell surface |
DPAGT1 15 |
p.L120L |
Markedly augments exon skipping, resulting in some skipped and infrequent non-skipped alleles |
CHRNA121 |
IVS3-8G>A |
Disrupts an ISS and enhances inclusion of a non-functional exon P3A, which disables expression of acetylcholine receptors at the cell surface |
DOK7 15 |
c.414C>T |
Deletion of the first 94 nucleotides of exon 4, and results a shift of in frame codons, predicted to be deleterious |
CHRNE11 |
ɛE154X |
Activation of a cryptic 3’ splice site in exon 6, results a PTC at the boundary of exons 6 and 7 |
DOK715 |
c.513C>G |
Generates a cryptic donor splice site, results an in-frame deletion of the last seven amino acid residues from exon 4 |
RAPSN16 |
IVS1-15C>A |
Generates a cryptic acceptor splice site. This causes |
CHRNE11 |
ɛEF157V |
Disrupts an ESE, and promotes skipping of exon 6. This results a PTC in exon 7 |
CHRNE11 |
ɛ553del7 |
Activates NASRE, which causes skipping of remote exon 6, results deletion of codons 148-187 |
Table 1 Representative mutations associated with aberrant splicing in CMS
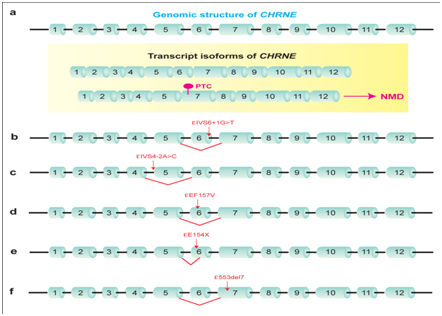
Figure 2 Representative mutations in CMS that disrupt cis-acting spicing elements in CHRNE encoding the acetylcholine receptor subunit.
In addition to mutations affecting the consensus splice sites, mutations disrupting the auxiliary splicing cis-elements (ISEs/ESEs and ISSs/ESSs) also show profound effects on splicing. These mutations may silence, enhance, or switch the activity (silencing to enhancing, and vice versa) of splicing regulatory cis-elements. ɛEF157V in CHRNEexon 6 causes exclusive skipping of exon 6 (Figure 2d).11 We found that ɛEF157V disrupts an ESE in exon 6, and compromises the binding of an SR protein, which subsequently induces skipping of exon 6 (manuscript in preparation). Additionally, we recently reported an interesting example of the detrimental effect of a single nucleotide mutation in a CMS patient.12 The catalytic subunits of acetyl cholinesterase (AChE) are anchored to the basal lamina of the NMJ using a collagen-like tail subunit (collagen Q, ColQ) encoded by ColQ. Mutations in ColQ cause endplate AChE deficiency.13 An A-to-G mutation predicting p.E415G in ColQ exon 16 identified in a patient with endplate AChE deficiency causes exclusive skipping of a constitutive exon 16.14 We found that the mutation disrupts binding of a splicing-enhancing RNA-binding protein, SRSF1, and de novo gains binding of a splicing-suppressing RNA-binding protein, hnRNP H (Figure 3a).12 Extensive analysis of spliceosome assembly demonstrated that the mutation compromised splicing of the downstream intron by impairing the formation of an early splicesome complex E. Further fractionation of constituents of E complex revealed that the mutation inhibits binding of U1 snRNP (prominently U1-70K) to the downstream 5’ splice site, which is facilitated by SRSF1 in the normal state. Therefore, a single nucleotide mutation causes the binding transition from SRSF1 to hnRNP H, and reverses the physiological splice-enhancing effect of the cis-element to the pathological splice-suppressing effect.
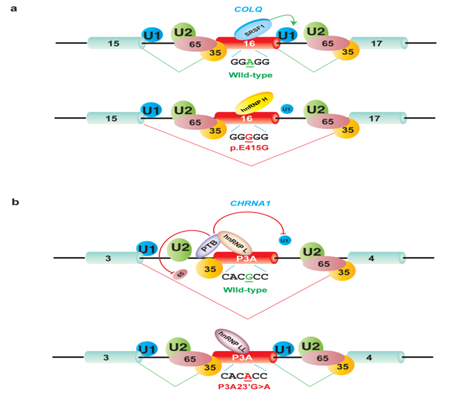
Figure 3 Opposite splicing regulation by antagonistic splicing trans-factors due to cis-acting point mutation associated with CMS. Large letters indicate functional binding of splicing trans-factors, whereas small letters represent compromised binding of splicing trans-factors. U1 denotes U1 snRNP; U2 denotes U2 snRNP; 65 denotes U2AF65; and 35 denotes U2AF35.Wild-type and mutant nucleotides are underlined.
Generation of a de novo splice site and activation of a cryptic splice site
The third and fourth classes of splicing defect are generation of a de novo splice site and activation of a cryptic splice site, which has been reported in several CMS (Table 1). A mutation sometimes generates AG or GT di-nucleotide within an intron or an exon. When the flanking nucleotides of AG or GT conform to the consensus splice site sequence and when conditions on the other cis-elements are permissible, the generated AG or GT dinucleotide is used as a de novo splicing donor or acceptor site. The generated splice site usually gains higher splice site strength over the native splice site, as measured by in silico splice site prediction tools. For example, c.513C>T in exon 4 of DOK7 generates a GT dinucleotide within exon 4, which is used as a novel splice donor site.15 Similarly, IVS1-15C>A in RAPSN de novo generates an AG dinucleotide in intron 1, and causes retention of 13 nucleotides of intron 1.16 An example of abnormal activation of a cryptic splice site is observed with ɛE154X in CHRNEexon 6, which disrupts an ESE and activates a cryptic 3’ splice site downstream of the mutant site (Figure 2e).11 Abnormal activation of a cryptic splice site also occurs due to inactivation or deletion of a native splice site. For example, IVS6-1G>C in CHRNE intron 6 inactivates the native 3’ splice site and abnormally activates a cryptic 3’ splice site in exon 7.17
HnRNP H, PTBP1, and hnRNP L cooperatively induce skipping of nonfunctional exon P3A of CHRNA1 encoding the acetylcholine receptor α subunit
Other interesting splicing mutations in CMS are relevant to cryptic exonization, in which a transposable element (TE) originating from short interspersed nuclear elements (SINEs) and mammalian interspersed repeats (MIRs) are changed to an exon in the course of evolution.18 Insertion of a TE into the host gene causes cryptic exonization of the inserted TE in most cases. In some cases, however, a second mutation on the inserted TE is required to activate cryptic exonization. An interesting example is exon P3A of CHRNA1. Exon P3A and its flanking intronic regions originate from exonization of the retroposed mammalian interspersed repeat element (MIR), which was inserted between exons 3 and 4 of CHRNA1.19 Inclusion of this in-frame exon P3A disables assembly of the AChR subunits. In human skeletal muscle, the P3A(-) and P3A(+) transcripts are generated in a 1:1 ratio.20 Exon P3A is present only in higher primates and in human. The reason why we have acquired the nonfunctional exon P3A in the course of evolution remains unsolved. In two patients with CMS, two distinct mutations caused exclusive inclusion of exon P3A.21,22 The first mutation (IVS3-8A>G) in intron 3 was at the eighth nucleotide preceding exon P3A (Table 1).22 Dissection of the underlying mechanisms revealed that the mutation disrupts an ISE, and compromises the binding of a cognate trans-factor, hnRNP H. This subsequently causes exclusive inclusion of exon P3A, which impedes the cell surface expression of AChR. As a result, neuromuscular signal transmission is compromised due to a reduction of AChR density at the patient endplate. The mechanisms in the second patient were more complicated. A missense mutation identified at the 23rd nucleotide of exon P3A (P3A23’G>A) causes aberrant inclusion of exon P3A.22 Mechanistic analysis revealed that the mutation gains a de novo binding affinity for a splicing enhancing factor, hnRNP LL, and competitively displaces binding of a splicing suppressing factor, hnRNP L (Figure 3b). The hnRNP L interacts with another splicing repressor PTBP1 through the proline-rich region and promotes binding of PTBP1to the poly pyrimidine tract upstream of exon P3A. Interaction of hnRNP L with PTBP1 inhibits association of U2AF65 and U1 snRNP with the upstream and downstream splice sites flanking exon P3A, respectively, which causes a defect in exon P3A definition and promotes exon skipping. In contrast, hnRNP LL lacks the proline-rich region and cannot interact with PTBP1. HnRNP LL thus antagonizes hnRNP L-mediated stabilization of PTBP1, and allows U2AF65 and U1 snRNP to associate with the upstream and downstream splice sites flanking exon P3A, which leads to abnormal inclusion of exon P3A.
Nonsense-associated altered splicing (NAS)
Generation of a PTC sometimes affects the consequence of splicing, and often causes skipping of a PTC-containing exon. This phenomenon is termed nonsense-associated altered splicing (NAS). NAS can be triggered through different mechanisms.23 The first mechanism is mediated by the nuclear scanning mechanism, which ensures translation of mature nascent mRNA lacking a PTC. A nonsense mutation interrupting the open reading frame can signal the splicing machinery to skip the affected exon. This type of selective exon skipping permits retention of the residual function of a protein instead of complete elimination of the coded protein by NMD. Second, when a PTC is in an alternative exon, an exon-included transcript is completely degraded by NMD, and an exon-skipped transcript becomes the only remaining transcript. Third, NAS is provoked when a nonsense mutation disrupts an ESE or compromises an essential splicing element in an RNA secondary structure. As far as we know, there is no NAS-provoking mutation in CMS.
Nonsense-associated altered splicing of a remote exon (NASRE)
A mutation may also drive skipping of a remote exon instead of the PTC-bearing exon. We reported a 7-bp deletion in CHRNEexon 7 in a CMS patient, which caused skipping of the preceding exon 6 carrying 101 nucleotides (Figure 2f).11 Due to the inherent weak strength of splice sites flanking exon 6, CHRNEnormally generates both exon 6-included and exon 6-skipped transcripts (Figure 2a). However, the exon 6-skipped transcript carries a PTC in exon 7, and is completely degraded by NMD. The 7-bp deletion in exon 7 resumes the open reading frame in the exon 6-skipped transcript, which makes the exon 6-skipped transcript resistant to NMD. In contrast, the exon 6-included full-length transcript is completely degraded by NMD due to the 7-bp frameshift mutation in exon 7. We termed this phenomenon as nonsense-associated altered splicing of a remote exon (NASRE). Skipping of a remote exon is also reported in other genes, such as SLC25A20,24 DBT,25 BTK,26 MLH1.27 The NASRE mechanism is able to explain aberrant skipping of a remote exon in these genes. NASRE is likely to be underestimated, because remote exons are seldom scrutinized in investigating genetic causes of human diseases.
HnRNP C, YB-1, and hnRNP L cooperate to make a Wnt-insensitive MUSK isoform
We recently reported an interesting splicing regulation of MUSK encoding MUSK, which plays an indispensable role in AChR clustering. MUSK exon 10 is alternatively spliced in human but not in mouse.28 MUSK mediates AChR clustering at the motor endplate and exon 10 encodes a frizzled-like cysteine-rich domain (Fz-CRD), which is essential for Wnt-mediated AChR clustering. We characterized splicing cis-elements in MUSK exon 10, where binding of hnRNP C promotes binding of YB-1 and hnRNP L to the immediate downstream sites and these three molecules cooperatively induce skipping of exon 10.28 Therefore, mutations in the identified splicing cis-elements in exon 10 potentially affect the function of MUSK by changing alternative splicing of exon 10, and can subsequently impede neuromuscular signal transmission. However, to the best of our knowledge, no mutations have been reported to date affecting these cis-elements in MUSK.
In silico programs to analyze splicing aberrations
A lot of web-based programs enable us to predict aberrant splicing due to a mutation and its underlying molecular mechanisms in silico, which should corroborated by in cellulo and in vitro analyses. Using these tools we can assess alternative splicing isoforms, splicing cis-elements, effects of mutations on splicing, predicting functional splicing regulatory motifs and their binding partner, strength of splice sites, RNA secondary structure, etc. Representative tools to predict splicing mutations at the 5’ and 3’ splice sites and the branch point are Human Splicing Finder,29 SD score,30 MaxEntScan,31 Analyzer Splice Tool (AST),31,32 DBASS5,33 DBASS3.34 Similarly, representative tools to predict splicing cis-elements and their associated RNA-binding proteins are SpliceAid,35 SpliceAid2,36 ESEfinder,37,38 Rescue-ESE,39,40 FAS-ESS,41 and SF map.42 The details of these tools as well as the other tools and databases are discussed in detail in our recent review article.43 Among these tools, we developed the SD score algorithm to predict the splicing effect of mutations affecting three nucleotides at the 3’ end of an exon and six nucleotides at the 5’ end of an intron.30 The SD-Score algorithm predicted aberrant splicing in 198 of 204 sites with a sensitivity of 97.1% and normal splicing in 36 of 38 sites with a specificity of 94.7%. An extensive simulation of all possible exonic mutations at positions -3, -2 and -1 at 189,249 5’ splice sites predicted aberration of pre-mRNA splicing in 37.8%, 88.8% and 96.8% of simulated mutations, respectively, which were all higher than we expected.
Antisense oligonucleotides correct splicing errors in CMS
Analyses of aberrant splicing in human diseases not only reveal the underlying maladies of splicing regulation in pathological conditions, but also provide an insight in splicing controls in physiological conditions.43,44 Therefore, we can exploit different biomedical strategies to reverse the aberrant splicing to a physiological state, which can hopefully be applied to clinical practice to develop rational therapy to treat splicing defects. Although splicing pathomechanisms in CMS have been dissected, a limited initiatives have been undertaken to develop rational therapies to treat CMS associated with aberrant splicing, compared to other inherited neuromuscular disorders such as spinal muscular atrophy (SMA) and Duchenne muscular dystrophy (DMD). In a recent study, antisense oligonucleotides (AONs) were employed to correct aberrant inclusion of exon P3A45 due to IVS3-8A>G and P3A23’G>A mutations that we addressed above.21,22 Three AON sequences were employed to sterically block the putative binding sequences for splicing factors necessary for exon recognition.45 They showed that AON complementary to the 5’ splice site of the exon was the most effective to induce exon skipping in the CHRNA1 minigene carrying wild-type sequence or harboring causative mutations. This study inspires the development of AON-based therapy for CMS patients.
Due to heterogeneity of clinical manifestations and the underlying mechanisms, a large number of CMS cases are predicted to be undiagnosed or misdiagnosed.2,46,47 CMS are not exceptional among Mendelian disorders from the viewpoint of germ line mutations. As in other Mendelian disorders, 15-20% of mutations affect splicing in CMS. We noticed that some missense and nonsense mutations, for which we have proven pathogenicity by analyzing mutant proteins, are splicing mutations and not missense or nonsense mutations. ɛE154X and ɛEF157V in CHRN11 and p.E415G in ColQ12,14 are these examples. Detailed dissections of aberrant splicing in CMS have disclosed yet unidentified mechanisms of alternative and constitutive splicing of genes expressed at the NMJ. Dissection of p.E415G in exon 16 of ColQ revealed that SRSF1 is essential for inclusion of constitutive exon 16.12 Dissection of IVS3-8G>A in intron 3 of CHRNA121 and P3A23’G>A in exon P3A of CHRNA122 disclosed that alternative skipping of exon P3A is mediated by hnRNP H, PTBP1, and hnRNP L. Although the analysis was not based on CMS mutations, we found that HnRNP C, YB-1, and hnRNP L cooperatively enhance skipping of MUSK exon 10 and make a Wnt-insensitive MUSK isoform.28 Splicing mutations prompted us to dissect the underlying mechanisms associated with the mutations. We, however, might have missed some mutations that were indeed splicing mutations but were recognized as missense or nonsense mutations. We should routinely screen for the effect of an identified mutation on splicing. Lack of expression of most NMJ molecules in blood cells and lack of available muscle specimens in most patients hinder us from analyzing the effect of the mutation on splicing in the native transcripts. Screening using a minigene is labor-intensive and is not realistic. To seek for an alternative to in cellulo and in vitro analyses, we analyzed hidden scenarios of the branch point,48 the 3’ splice site,49 and the 5’ splice site50 by exploiting in silico analysis methods. Tools to predict RNA-binding proteins for specific cis-elements are also available online,29,42 and further development of such tools will facilitate identification of splicing mutations and their underlying mechanisms.
None.
Author declares that there is no conflict of interest.

©2015 Rahman, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.