Journal of
eISSN: 2373-4469


Review Article Volume 2 Issue 1
Division of Neurogenetics, Center for Neurological Diseases and Cancer, Nagoya University Graduate School of Medicine, Japan
Correspondence: Kinji Ohno, Division of Neurogenetics, Center for Neurological Diseases and Cancer, Nagoya University Graduate School of Medicine, 65 Tsurumai, Showa-ku, Nagoya, Aichi 466-8550, Japan, Tel +81 52 744 2446, Fax +81 52 744 2449
Received: January 09, 2015 | Published: January 21, 2015
Citation: Rahman MA, Nasrin F, Masuda A, et al. Decoding abnormal splicing code in human diseases. J Investig Genomics. 2015;2(1):6-23. DOI: 10.15406/jig.2015.02.00016
RNA splicing is an intricate process in humans and higher metazoans. Splicing is regulated through multifaceted coordinated factors, such as cis-acting splicing code and RNA-binding splicing trans-factors that associate or compete with ribonucleoproteins (RNPs). Individual cis-acting splicing code and their functional coordination with cognate splicing trans-factors still remain elusive mostgenes, because these code are comprised of highly degenerative short sequence motifs and multiple splicing trans-factors can recognize an identical motif. In addition, a specific splicing motif functions differentially in different genes, which is determined by additional factors such as neighboring sequence context, cell types and association/competition with other splicing trans-factors. Genetic and cellular alterations compromising the fidelity of splicing processes provoke many human diseases. Analyses of abnormal splicing code in human diseases not only uncover the underlying maladies of splicing regulations in pathological conditions, but also allow us to gain insight into splicing mechanisms in physiological conditions. This review introduces accumulating knowledge of numerous modes of splicing aberrations and provides critical information to understand the underlying patho mechanisms of human diseases, which hopefully leads to development of rational therapies.
Keywords: alternative splicing, cis-acting splicing code, splicing trans-factor, spliceosome, mutation, aberrant splicing, neurodegenerative diseases, cancers
RNPs, ribonucleoproteins; snRNPs, small nuclear ribonucleoproteins; snRNA, small nuclear RNA; BP, branch point; PPT, poly pyrimidine Tract; ISE, intronic splicing enhancer; ESE, exonic splicing enhancer; ISS, intronic splicing silencer; ESS, exonic splicing silencer; SR Proteins, serine/arginine-rich proteins; RRM, RNA-recognition motif; RS Domains, arginine and serine rich domains; hnRNPs, heterogeneous nuclear ribonucleoproteins; ARE, au-rich element; NMD, nonsense mediated decay; AChR, acetylcholine receptor; Fz-CRD, frizzled-like cysteine-rich domain; TE, transposable elements; SINEs, short interspersed nuclear Elements; MIRs, mammalian interspersed repeats; FCMD, fukuyama congenital muscular dystrophy; CMS, congenital myasthenic syndrome; SMA, spinal muscular atrophy; ALS, amyotrophic lateral sclerosis; CLIP, cross-linked immuno precipitation; FTD, fronto temporal dementia; NMJ, neuromuscular junction; AD, alzheimer’s disease; DCM, dilated cardiomyopathy; PAR-CLIP, photoactivatable ribonucleoside–enhanced crosslinking and immuno precipitation; NAS, nonsense-associated altered splicing; NASRE, nmd-associated skipping of a remote exon; PTC, premature termination codon; MD, myotonic dystrophy; NSCLC, non-small cell lung cancers; HPT-JT, hyper parathyroidism-jaw tumor syndrome; FIHP, familial isolated primary hyper parathyroidism; FED, fish-eye disease; LQTS, long qt syndrome; CCA, congenital contractural arachnodactyly; IGHD, isolated growth hormone deficiency; FHL3, familial hemophagocytic lymphohistiocytosis; PMD, pelizaeus–merzbacher disease; GHI, growth-hormone insensitivity; RP, retinitis pigmentosa; FTLD, fronto temporal lobar degeneration; ASD, autism spectrum disorders
Humans and other higher metazoans acquired regulated diversity in their genes by inserting multiple noncoding introns into a coding region. At a cost of excising noncoding introns every time a gene is transcribed, we expanded proteome diversity by alternative splicing without increasing the NUMBer of genes. About 95% of human multi-exon genes are estimated to undergo alternative splicing.1,2 To support physiological and cellular demand alternative splicing is fine-tuned in tissue-specific, developmental stage-specific and gender-specific manners. It is also modulated in response to external stimuli or intracellular signals. We humans utilize the most complex alternative splicing events. However, with the increase of splicing complexity, splicing processes became readily susceptible to mis regulation, which potentially affects cellular physiology and gives rise to various human diseases. Scrutinized dissection of abnormal splicing mechanisms in human diseases ironically enable us to disclose yet unidentified splicing scenarios in physiological conditions. In this review, we present some examples in which the mechanisms of missplicing are well dissected and implicated in understanding physiological splicing regulations.
Splicing machinery and splicing code
Splicing of pre-mRNA is carried out in nucleus by spliceosome, which is a macromolecular complex composed of RNA and proteins. Five small nuclear ribonucleoproteins (snRNPs) and multiple proteins (>100) cooperate in spliceosome to catalyze the splicing reaction. Each snRNP is composed of a single uridine-rich small nuclear RNA (snRNA) and multiple proteins. Spliceosome-mediated splicing can be achieved by two steps: recognition of intron/exon boundary and catalysis of the transesterification reaction to excise out an intron followed by joining two exons. Recognition of intron/exon boundary is guided by essential splicing cis-elements close to either end of an intron, termed as consensus splice site sequences (Figure 1a). These include 5’ splice site, a branch point (BP), a poly pyrimidine tract (PPT), and 3’ splice site. In humans and metazoans, these consensus splice site sequences are highly degenerative, which partly compromises appropriate recognition and binding of essential trans-acting factors. As a consequence, multiple auxiliary trans-acting factors need to co-operate to form spliceosome and to favor folding of nuclear pre-mRNA to commit splicing. The assembly of spliceosome is further regulated by auxiliary splicing cis-elements either in a positive or negative manner (Figure 1b). Positively modulating cis-elements are termed as intronic/exonic splicing enhancers (ISEs/ESEs), whereas negatively modulating cis-elements are termed as intronic/exonic splicing silencers (ISSs/ESSs). Most of these auxiliary cis-elements function by binding to cognate trans-factors (Table 1), whereas some cis-elements function by forming secondary structures. The majority of splicing trans-factors for ESE are serine/arginine-rich (SR) proteins, which function as either an essential or regulatory factor.3,4 SR proteins can modulate several steps of spliceosome assembly through protein-protein and protein-RNA interaction.5,6 They possess one or two RNA-recognition motif (RRM) at the N-terminal end and arginine and serine residues (RS domains) at the C-terminal end. The majority of splicing trans-factors for splicing silencer elements (ISSs/ESSs) are heterogeneous nuclear ribonucleoproteins (hnRNPs).7 Members of this family usually contain an RRM-type and KH-type RNA-binding domain along with other auxiliary domains to mediate protein-protein interaction. ISEs are less well characterized compared to the other cis-elements. Recent analyses on ISEs suggest that hnRNP F, hnRNP H, NOVA1, NOVA2, FOX1, and FOX2 are candidate factors for ISEs.8‒11
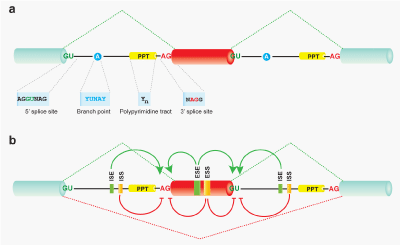
Figure 1 cis-acting splicing code
(a) Schematic of essential splicing cis-elements: invariant GU and AG dinucleotides constituting the 5’ and 3’ splice sites, respectively, at the ends of an intron; the branch point (BP, ‘A’ is observed at 92.3%); and the polypyrimidine tract (PPT). The consensus sequences of these elements are shown below (Y = C/U, R = G/A, N = any nucleotide).
(b) Schematic of auxiliary splicing cis-elements, which can influence alternative splicing. Based on location and functional activity, these elements can be categorized into intronic/exonic splicing enhancers (ISEs/ESEs) and intronic/exonic splicing silencers (ISSs/ESSs). Recognition of splice sites are promoted by enhancer elements and repressed by silencer elements. In addition, enhancers can antagonize the activity of silencers, and vice versa. Exon inclusion or skipping is finely regulated by the relative strength of these influential elements, which are mostly determined by binding of cognate splicing trans-factors. Activities of splicing trans-factors are also regulated by developmental stage-specific and tissue-specific expression, as well as by phosphorylation and post translational modifications. Therefore, fine-tuned coordination of splicing cis-elements and spatiotemporal expressions of splicing trans-factors is critical to attain the transcriptome and proteome diversities that we human have acquired in the course of evolution.
Protein |
Binding sequence |
SR proteins |
|
SRSF1 (SF2/ASF)106‒108 |
SRSASGA, RGAAGAAC, AGGACRRAGC, GGAGA |
SRSF2 (SC35)107‒110 |
UGCNGYY, GRYYCSYR, AGSAGAGUA, GUUCGAGUA, GA-rich sequence |
CCUCGUCC, GCUCCUCUUCC, WCWWC |
|
GAAGGA, GAUGA, AAGAA, GAAGA, AGAAG, GAAAA |
|
ACDGS, UGGGAGCRGUYRGCUCGY |
|
SRSF6 (SRp55)106 |
USCGKM |
SRSF7 (9G8)112 |
(GAC)n |
HnRNPs |
|
UAGGG(A/U), UAGG |
|
hnRNP A2/B1117‒119 |
(UUAGGG)n, UAG, UAGRGA |
Poly-U |
|
G-rich sequences |
|
G-rich sequences |
|
CU-rich sequences, UYUYU |
|
CA-repeat or C/A-rich sequences |
|
CA-repeat or C/A-rich sequences |
|
Other splicing regulatory proteins |
|
U/G-rich sequences |
|
ETR3 (CELF2)130 |
U/G-rich sequences |
CELF4131 |
U/G-rich sequences |
ACUAAY |
|
YGCU(U/G)Y |
|
FOX1136 |
(U)GCAUG |
FOX2136 |
(U)GCAUG |
YCAY |
|
YCAY |
|
TIA1139 |
U-rich sequences |
TIAR139 |
U-rich sequences |
nPTB140 |
CU-rich sequences |
CU-rich sequences |
|
RBM5143‒145 |
CU-rich sequences, GA-rich sequences, poly(G), ANGUAA |
RBM6145 |
CUCUGAA |
RBM10145 |
ANGUAA, CUCUGAA, poly(G), |
RBM20146 |
UCUU |
RBM35a (ESRP1)147 |
GU-rich sequence |
RBM35b (ESRP2)147 |
GU-rich sequence |
Table 1 cis-acting RNA motifs recognized by representative splicing regulatory trans-acting proteins
R (purine)=A/G, Y (pyrimidine)=C/U, W (weak hydrogen bonds)=A/U, S (strong hydrogen bonds)=C/G, K(keto in a large groove)=G/U, and M(amino in a large groove)=A/C
Spliceosome assembly starts with the recognition of the 5’ splice site by U1 snRNP, the BP by SF1, and the PPT as well as 3’ terminal AG by U2AF heterodimer (U2AF65 and U2AF35, respectively) (Figure 2). This is an ATP-independent initial assembly step termed as an E complex. Initial spliceosome assembly is usually formed across a single intron in a two-exon gene or short introns in multi-exon genes (termed as an intron-defined E complex), whereas it is formed across an exon flanked by long introns in higher metazoans (termed as exon-defined E complex).12‒14 The ATP-independent E complex is then transformed to ATP-dependent spliceosome A complex, where SF1 is replaced by U2 snRNP at BP. Subsequent recruitment of U4/U6-U5 snRNPs leads to the formation of B complex. Through extensive remodeling and conformational changes, an active spliceosome complex called C complex is formed by replacing U1 and U4 snRNPs, which subsequently catalyzes splicing. In most cases, splicing activators or repressors function by modulating the early spliceosome assembly at the stage of E complex or A complex. Therefore, the ultimate splicing consequence (constitutive splicing or alternative splicing) is determined by complex cis-acting splicing code, their cognate RNA-binding trans-factors and an immense network of their interactions.
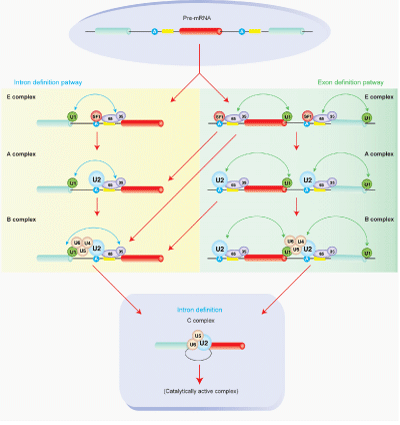
Figure 2 Schematic of major spliceosome assembly on pre-mRNA. The recognition of exon and concomitant intron removal is preceded through the coordinated assembly of spliceosome either across an intron (termed as the intron-definition pathway) or across an exon (termed as the exon-definition pathway). The intron-definition pathway is favored in pre-mRNAs containing a single or short intron, whereas the exon-definition pathway is favored in pre-mRNAs containing a long intron as in metazoans. The progression of spliceosome assembly starts with the recognition of the 5’ splice sites by U1 snRNP, the BP by SF1, and the PPT as well as 3’ terminal AG by U2AF heterodimer consisting of U2AF65 (65) and U2AF35 (35). This is an ATP-independent initial assembly step forming the E complex. The ATP-independent E complex is then transformed to the ATP-dependent spliceosome A complex, where SF1 is replaced by U2 snRNP at BP. Subsequent recruitment of U4/U6-U5 snRNPs leads to the formation of the B complex. Through extensive remodeling and conformational changes, the active spliceosome C complex is formed by replacing U1 and U4 snRNPs, which subsequently catalyzes splicing. Various lines of evidence demonstrate that transition of spliceosome from exon-definition to intron-definition occurs in the course of splicing of a single exon, which is determined by multiple factors such as the splice site strength, the intron length, regulatory trans-factors, etc.12-14 Both the exon-definition and intron-definition pathways merge into formation of the final catalytic C complex formed across an intron for accurate intron removal.
Pre-mRNA splicing defects cause human diseases
To support rapidly changing cellular processes, splicing should be rapid and precise. Therefore, correct complement of RNA and proteins in the right cell at the right time is indispensable for strictly regulated biogenesis of ribonucleoprotein complexes (RNPs). Mutations affecting cis-elements, mutations in trans-acting factors and over/under-expressions of trans-acting factors potentially impair formation of functional spliceosomes. These result in deleterious consequences to cells and underlie a variety of human diseases.
cis-scting splicing defects associated with human diseases
About 15 to 20% of human genetic diseases are caused by mutations in the consensus splice site sequences or in the auxiliary splicing cis-elements.15 cis-acting mutations can interrupt the recognition of both constitutive and alternative splice sites by their cognate trans-acting factors. When a splice site is affected, the aberrantly spliced gene may lack an essential cis-element for mRNA degradation such as AU-rich element (ARE).16 Similarly, a protein encoded by the aberrantly spliced gene may lack essential domains and signals. Generation of a premature termination codon can give rise to a truncated protein with abnormal functions or activate the nonsense mediated decay (NMD) pathway. In addition, when an alternative splice site is affected, an abnormal change in the ratio of alternative iso forms may compromise cellular processes.
Defects in essential splicing cis-elements: A lot of human diseases are due to mutations in consensus splice sites (see some examples in Table 2). The consensus of the 5’ splice site is “CAG/GUAAGU” where “/” denotes an exon-intron boundary.17,18 This site is recognized by U1 snRNP. In humans and other higher eukaryotes, BP sequences are highly degenerative, whereas yeast has a strictly conserved BP sequence of ‘UACUAAC’.19 To elucidate a consensus sequence of human BP, we analyzed 367 lariat RT-PCR clones arising from 52 introns of 20 human housekeeping genes and disclosed that the consensus human BP sequence is “yUnAy” (y = C/U and n= any nucleotide).20 The fourth nucleotide “A” is the branch point (position +0) and is conserved in 92.3% of the clones. The “U” at position -2 is conserved in 74.6%. Collation of 46 experimentally confirmed BPs in previous reports also gave rise to our BP consensus sequence of “yUnAy”. Extensive analyses of human BPs using RNA-seq data similarly revealed that the consensus sequence is “UnAy”.21,22
Gene |
Mutation |
Consequence |
Disease |
Disruption of consensus splice sites |
|||
5’ splice site-disrupting mutations |
|||
HRPT2148 |
IVS2+1G>C |
Exon 2 skipping |
Hyperparathyroidism-jaw tumor syndrome (HPT-JT) |
HRPT2149 |
IVS1+1G>A |
Partial deletion |
Familial isolated primary hyperparathyroidism (FIHP) |
CHRNE150 |
IVS7+2T>C |
Intron 7 retention |
Congenital myasthenic syndrome (CMS) |
Branch point-disrupting mutations |
|||
LCAT151 |
IVS4-22T>C |
Intron 4 retention |
Fish-eye disease (FED) |
KCNH2152 |
IVS9-28A/G |
Intron 9 retention |
Long QT syndrome (LQTS) |
FBN2153 |
IVS30-26T>G |
Exon 31 skipping |
Congenital contractural arachnodactyly (CCA) |
Polypyrimidine tract-disrupting mutations |
|||
FANCA154 |
c.710-5T>C |
Exon 8 skipping |
Fanconi anemia |
RB1155 |
IVS8-10T>C |
Exon 9 skipping |
Retinoblastoma |
DFNA5156 |
IVS7-DCTT |
Exon 8 skipping |
Nonsyndromic hearing impairment |
3’ splice site-disrupting mutations |
|||
HPRT2157 |
IVS2-1G>A |
Exon 3 skipping |
HPT-JT |
CSPG2158 |
IVS7-2A>G |
Activation of a cryptic splice site |
Wagner syndrome |
CHRNE159 |
IVS6-1G>C |
Activation of a cryptic splice site |
CMS |
Disruption of auxiliary splicing cis-elements |
|||
ESE-disrupting mutations |
|||
COLQ 160 |
p.E415G |
Exon 16 skipping |
CMS |
CHRNE76 |
p.EF157V |
Exon 6 skipping |
CMS |
BRCA188 |
p.G5199T |
Exon 18 skipping |
Breast and ovarian cancer |
ESS-disrupting mutations |
|||
CHRNA 32 |
P3A23’G>A |
Exon P3A inclusion |
CMS |
MAPT161 |
p.N269N |
Exon 10 inclusion |
Neurodegenerative tauopathies |
PTPRC162 |
p.C77G |
Skipping of multiple exons |
Multiple sclerosis |
ISE-disrupting mutations |
|||
GH1163 |
IVS3+28G>A |
Exon 3 skipping |
Isolated growth hormone deficiency (IGHD II) |
UNC13D164 |
IVS1+525G>T |
Retention of an intronic segment |
Familial hemophagocytic lymphohistiocytosis (FHL3) |
PLP1165 |
IVS3D28-46 |
Alternative 5’ splice site selection |
Pelizaeus–Merzbacher disease (PMD) |
ISS-disrupting mutations |
|||
CHRNA127 |
IVS3-8G>A |
Exon P3A inclusion |
CMS |
ATM166 |
4 bp deletion in intron 20 |
Activation of a cryptic exon |
Ataxia-telangiectasia |
GHR 167 |
c.618+1800A>G |
Activation of a cryptic exon |
Inherited growth-hormone insensitivity (GHI) |
Table 2 Representative cis-acting defects associated with aberrant splicing
A high degree of degeneracy of human BP sequences suggests that recognition of human BP is likely to be cooperated along with downstream PPT where U2AF65 binds and possibly the invariant AG dinucleotide at the 3’ splice site where U2AF35 binds. In PPT, uridines are preferred over cytidines.23,24 PPT with eleven continuous uridines is highly competent and the position of such PPT is not critical.23 On the other hand, PPTs with only five or six uridines are required to be located close to the 3’ AG for efficient splicing.23 It is interesting to note that introns carrying a long stretch of PPT favors the binding of U2AF65 so strong that binding of U2AF35 to 3’ splice site becomes dispensable, which is called “AG-independent 3’ splice site”. In contrast, when the PPT is short, binding of U2AF35 becomes indispensable for splicing, which is called “AG-dependent 3’ splice site”. Again, the proximity between BP and the 3’ splice site is sometimes a critical factor for splice site selection.24 Therefore, a synergistic co-regulatory process decides the efficient recognition of the intron/exon boundary at the 3’ splice site. The first nucleotide of an exon (E+1 ) is also sometimes correlated with splicing. Mutations at E+1 is also prevalent in human diseases, but is less analyzed compared to the invariant ‘AG’ at the 3’ splice site. We dissected the splicing effects of mutations at E+1 , and found that an E+1 mutation causes aberrant splicing at an AG-dependent splice site and normal splicing at an AG-independent splice site.25 We reported five mutations at E+1 , which caused aberrant splicing. In the course of our analysis, we detected that in the human genome, T is preferentially selected at exonic positions +3, +4 and +5, which was partly consistent with a previously reported in vitro SELEX study demonstrating that U2AF35 can bind up to 12 nucleotides downstream from the intron-exon boundary.26 In the U2AF35-selected RNA pool, enrichment of T at exonic positions +3, +4 and +5 were indeed evident. Although, splicing mutations affecting T nucleotide at these exonic positions have not been reported so far, but we assume that some of these mutations confer susceptibility to aberrant splicing.
Defects in auxiliary splicing cis-elements: In addition to mutations affecting the consensus splice sites, mutations affecting auxiliary cis-elements (ISEs/ESEs and ISSs/ESSs) also have profound effects on aberrant splicing. These mutations may silence, enhance or switch the activity (silencing to enhancing, and vice versa) of splicing regulatory cis-elements. We have summarized representative examples of aberrant splicing in human diseases due to the defect in auxiliary splicing cis-elements in Table 2. A remarkable example of the detrimental effect of a single nucleotide mutation was evident in CHRNA1 encoding the muscle nicotinic acetylcholine receptor alpha subunit. CHRNA1 harbors an inframe alternatively spliced exon P3A and inclusion of this exon disables assembly of the acetylcholine receptor (AChR) subunits and subsequently prevents their expression on the cell surface. In two different patients suffering from congenital myasthenic syndrome, a point mutation was identified in each patient with striking physiological consequences (Figure 3a) (Figure 3b). The first mutation (IVS3-8A>G) in intron 3 was identified at the eighth nucleotide preceding exon P3A (Figure 3a).27 Detailed analysis demonstrated that the mutation disrupts an ISE and compromises the binding of a cognate trans-factor, hnRNP H. This subsequently causes exclusive inclusion of exon P3A, which impedes the cell surface expression of AChR. As a result, neuromuscular signal transmission is compromised due to a reduction of AChR density at the patient end plate. The mechanisms in the second patient were more complicated (Figure 3b). A missense mutation identified at the 23rd nucleotide of exon P3A (P3A23’G>A) causes aberrant inclusion of exon P3A and subsequently compromises neuromuscular signal transmission at the patient end plate. Mechanistic analysis revealed that the mutation gains a de novo binding affinity for a splicing enhancing factor, hnRNP LL, and displaces binding of a splicing suppressing factor, hnRNP L. The hnRNP L interacts with another splicing repressor PTB through the proline-rich region and promotes PTB binding to the polypyrimidine tract upstream of exon P3A. Interaction of hnRNP L with PTB inhibits association of U2AF65 and U1 snRNP with the upstream and downstream splice sites flankingexon P3A respectively, which causes a defect in exon P3A definition and promotes exon skipping. In contrast, hnRNP LL lacks the proline-rich region and cannot interact with PTB. HnRNP LL thus antagonizes hnRNP L-mediated stabilization of PTB and U2AF65 and U1 snRNP are able to associate with the upstream and downstream splice sites flanking exon P3A, which leads to inclusion of exon P3A.
Binding of antagonizing splicing trans-factors to an identical site is observed in SMN1 and SMN2 (Figure 3c). SMN1 and SMN2 are highly homologous paralogs with only a single nucleotide substitution. SMN1 and SMN2 carry C and T, respectively at the 6th nucleotide of exon 7. Splicing of SMN1 exon 7 is enhanced by SRSF1.28,29 A C-to-T substitution at the 6th nucleotide of exon 7 in SMN2 gains binding of a splicing-suppressing hnRNP A1.30,31 In addition, the C-to-T substitution may28,29 or may not30,31 abolish binding of SRSF1. In contrast to hnRNPs L and LL for CHRNA1,32 however SRSF1 and hnRNP A1 do not compete for binding to an identical site. Although no splicing mutations have been reported, we recently reported that exon 10 of MUSK encoding the muscle-specific receptor tyrosine kinase, MuSK, is alternatively spliced in human but not in mouse.33 MuSK mediates AChR clustering at the motor endplate and exon 10 encodes a frizzled-like cysteine-rich domain (Fz-CRD), which is essential for Wnt-mediated AChR clustering. In MUSK exon 10, binding of hnRNP C promotes binding of YB-1 and hnRNP L to the immediate downstream sites and these three molecules cooperatively enhance skipping of exon 10. As the splicing cis-elements are within exon 10, mutations affecting MUSK potentially cause exon skipping, but no mutations reported to date affect these elements.
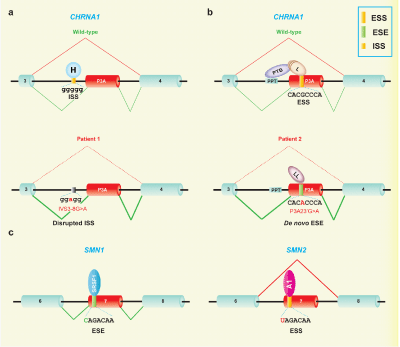
Figure 3 Splicing consequences due to disruption of cis-acting splicing code.
(a) In a patient with congenital myasthenic syndrome (CMS), a mutation in intron 3 (IVS3-8G>A) of CHRNA1 disrupts an intronic splicing silencer (ISS) by compromising the binding of its cognate factor, hnRNP H (H).27 This causes an exclusive inclusion of a nonfunctional exon P3A, which disrupts the assembly of acetylcholine receptor (AChR) on the cell surface, thereby compromises neuromuscular signal transmission at the patient endplate.
(b) In a second patient with CMS, a missense mutation in exon P3A (P3A23’G>A) of CHRNA1 disrupts binding of a splicing suppressing RNA-binding protein, hnRNP L (L), and de novo generates a binding affinity for a splicing enhancing RNA-binding protein, hnRNP LL (LL).32 HnRNP L normally stabilizes binding of another splicing repressor, PTB to the upstream polypyrimidine tract (PPT) of P3A, which is disrupted due to the mutation. As a result, aberrant inclusion of exon P3A is observed with subsequent maladies at the patient endplate.
(c) Spinal muscular atrophy (SMA) is characterized by degeneration of spinal motor neurons and is caused by loss-of-function mutations in the survival of motor neuron 1 gene (SMN1). SMN1 and SMN2 are nearly identical paralogues, which carry C and T, respectively, at the 6th nucleotide of exon 7. Inclusion of SMN1 exon 7 is enhanced by SRSF1 through an ESE [28,29]. A C-to-T substitution at the 6th nucleotide of exon 7 inactivates the binding of splicing enhancing SRSF128,29 and gains binding of splicing-suppressing hnRNP A1(A1).30,31 As a result, a functionally compromised exon 7-skipped isoform (SMN7) is produced, which cannot compensate for the loss of SMN1 in SMA, leading to progressive degeneration of spinal motor neurons.
Defects in RNA secondary structure: In addition to disruption of consensus and auxiliary splicing cis-elements, mutations may affect the RNA secondary structure, which also have a critical role in pre-mRNA splicing.34‒37 The most commonly shared feature is the presence of structural elements that interfere with the accessibility of essential splicing factors to the cognate cis-elements. In some cases, structural constraints outside the essential cis-elements can also affect splicing process indirectly by modulating the relative distance between splice sites, thereby generating variability in splice site recognition efficiency. In addition, RNA structural features have also been documented to affect the accessibility of auxiliary splicing cis-elements (ESEs/ISEs and ESSs/ISSs). Therefore, genetic defects can potentially affect splicing by altering the RNA secondary structures, which can give raise to devastating pathological abnormalities in humans. The RNA secondary structure and its association with human disease has been deeply dissected in splicing of human tau exon 10.38‒41 Mutations in the tau gene (MAPT) are associated with frontotemporal dementia and Parkinsonism. A cluster of mutations is located at an RNA stem-loop structure at the 3’ end of exon 10. This stem-loop structure restricts the accessibility of U1 snRNP to the 5’ splice site and thereby critically regulates alternative splicing of tau exon 10 to attain a physiological ratio of exon 10-skipped and included transcripts. Mutations in this region destabilize the stem-loop structure and subsequently increase the splice site usage due to an enhanced recognition by U1 snRNP. As a result, physiological balance of the two transcripts is disrupted owing to the abnormal production of an exon 10-included tau isoform, which leads to tau aggregates in neuronal and glial cells.
Aberrant activation of cryptic splice site: Another class of splicing defect is the activation of a cryptic splice site, which causes retention of a segment within an intron. Inclusion of an intronic segment in spliced mRNA often causes disruption of the open reading frame or generation of a premature termination codon. Abnormal activation of a cryptic splice site has been reported in many human diseases.42 Activation of a cryptic splice site is mostly due to a mutation generating a de novo splicing donor or acceptor site within an intron. The generated splice site usually possesses higher splice site strength over the native splice site, as measured by in Silico web programs. Alternatively, inactivation or deletion of a native splice site can also potentiate the selection of a cryptic splice site. A rare mechanism was identified in Duchenne muscular dystrophy, where genomic inversion causes activation of cryptic splice sites.43
Cryptic exonization: Besides cryptic splice site activation, a closely relevant event is cryptic exonization, which is mediated by transposable elements (TE), originating from short interspersed nuclear elements (SINEs) and mammalian interspersed repeats (MIRs).44 In most cases, insertion of TE into the host gene directly causes cryptic exonization of the inserted TE. In some cases, a second mutation on the inserted TE is required to activate cryptic exonization. An example of direct exonization is the fukutin gene (FKTN), where cryptic exonization of a TE inserted at the 3’ UTR of FKTN produces a transcript isoform encoding a nonfunctional protein with an altered C-terminus, leading to Fukuyama congenital muscular dystrophy (FCMD).45 Another interesting example is exon P3A of CHRNA1 gene, which we discussed in the previous section. Exon P3A and its flanking intronic regions have been arisen from exonization of the retroposed mammalian interspersed repeat element (MIR).46 Inclusion of this inframe exon P3A disables assembly of the AChR subunits. In human skeletal muscle, the P3A(-) and P3A(+) transcripts are generated in a 1:1 ratio.47 Acquisition of exon P3A is predicted to be detrimental for human, because exclusive inclusion of exon P3A would compromise neuromuscular signal transmission by reducing AChR expression at the endplate. Indeed in two individual cases, mutations causing exclusive inclusion of exon P3A give rise to congenital myasthenic syndrome (CMS), which is characterized by abnormal muscle fatigue, muscle weakness, amyotrophy and sometimes minor facial anomalies (Figure 3).27,32
Trans-acting splicing defects causing human diseases
Compared to cis-acting splicing defects, trans-acting defects can potentially exert a more detrimental consequence, because multiple target genes can be affected by a defect of a single trans-factor. The affected trans-factors can be either an essential constituent of splicing machinery or an auxiliary factor modulating alternative splicing. The defect can be either a genetic mutation in a trans-factor itself or nongenetic aberration affecting the fidelity of recruitment of a trans-factor to spliceosome. Both can lead to altered splicing efficiency of both constitutive and alternative exons. Nongenetic aberrations of trans-factors include abnormal expression, post-translational modification, and subcellular localization of trans-factors (Figure 4).
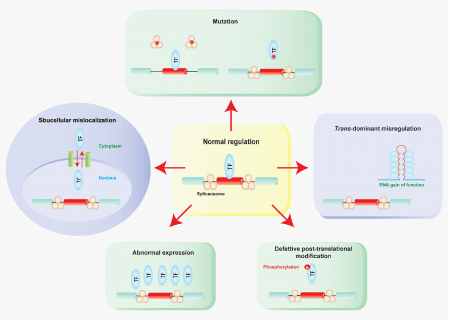
Figure 4 Models of trans-acting splicing defects. Schematic summary of different mode of splicing aberrations due to defects in trans-acting splicing factors including mutations in both essential and auxiliary trans-factors, subcellular mislocalization, abnormal expression, defective post-translational modification, and trans-dominant splicing defects. TF indicates a trans-factor.
Defects in essential splicing factors: Mutations in the core spliceosome components have been identified as a potential cause of many human splicing diseases in recent years (Table 3). A striking example is myelodysplasia characterized by deregulated dysplastic blood cell production with predisposition to acute myeloid leukemia. In a study of whole-exome sequencing of 29 myelodysplasia specimens, mutations involving multiple components of spliceosome have been demonstrated as the cause of pathogenesis.48 The mutations are mostly in SF3B1, SRSF2, U2AF35, and ZRSR2 and less frequently in SF3A1, SF1, U2AF65 and PRPF40B. These mutated trans-factors are confined to spliceosome E complex and A complex, suggesting that compromised function of spliceosome E/A complex is a potential cause of myelodysplasia. Another interesting example is autosomal dominant forms of retinitis pigmentosa (Table 3), which is caused by the loss of retinal rod photoreceptor cells due to mutations in the essential splicing factors of PRPF3, PRPF8, PRPF31 and RP9.49 A defect in the biogenesis of essential spliceosome components is also exemplified in spinal muscular atrophy (SMA), which is caused by the deficiency of SMN leading to motor neuron degeneration. SMN complex is essential for biogenesis of snRNPs and SMN-deficiency perturbs the stoichiometry of snRNAs, which causes wide-ranging pre-mRNA splicing defects in numerous genes.50
Gene |
Defect |
Consequence |
Disease |
Defects in essential trans-factors in spliceosome |
|||
SF3B1, |
Mutation |
Predicted to affect spliceosome |
Myelodysplasia |
SF3A1, |
|||
U2AF35, U2AF65, |
|||
ZRSR2, |
|||
SF1, |
|||
PRPF40B48 |
|||
PRPF31, |
Mutation |
Predicted to affect spliceosome |
Retinitis pigmentosa (RP) |
PRPF8, |
assembly by compromising function of tri-snRNPs (U4/U6-U5) |
||
PRPF3, |
|||
PAP149 |
|||
Defects in auxiliary splicing trans-factors |
|||
TARDBP |
Mutation |
Aberrant splicing regulations due to |
Amyotrophic lateral sclerosis (ALS), frontotemporal lobar degeneration (FTLD), and Alzheimer’s disease (AD) |
Mutation |
Aberrant splicing regulations due to |
ALS |
|
Altered expression |
Altered splicing and transcriptional networks in neurons |
Autism spectrum disorders (ASD) |
|
hnRNPs A2B1 and A1173 |
Mutation |
Altered dynamics of ribonucleoprotein granule assembly and predicted to affect splicing |
Multisystem proteinopathy, ALS |
HMGA1a67 |
Induced expression |
Aberrant skipping of exon 5 of PS2 |
Sporadic AD |
Abnormal expression |
Aberrant splicing of exon 10 of tau gene |
Tauopathies in neurodegenerative disorders, AD |
|
Auto antibodies against NOVA-1 |
Aberrant splicing of NOVA-1-regulated pre-mRNAs in neurons |
Paraneoplastic syndrome |
|
hnRNP L104 |
Phosphorylation |
Aberrant splicing of CASP9 |
Tumorigenesis in non-small cell lung cancer |
QKI103 |
Reduced expression |
Aberrant splicing of NUMB gene |
Lung cancer |
Abnormal expression |
Aberrant splicing of SRSF1-regulated genes |
Cancers |
|
Abnormal expression |
Aberrant splicing of SRSF3-regulated genes |
Cancers |
|
Table 3 trans-acting defects associated with aberrant splicing
First, TDP-43 regulates alternative splicing. For example, TDP-43 displays a substantial binding affinity for the microsatellite region (UG)nin intron 8 of CFTR(a gene mutated in cystic fibrosis) and subsequently induces skipping of exon 9.54 This causes generation of a functionally compromised exon 9-skipped protein isoform, which was previously demonstrated to be associated with cystic fibrosis and congenital bilateral absence of the vas deferens.55,56 To further address the nuclear dysfunction, in vivo RNA targets of TDP-43 have been extensively identified by cross-linked immunoprecipitation (CLIP)-seq.57,58 CLIP-seq revealed that TDP-43 preferentially regulates splicing of mRNAs that are important for brain development and synaptic function. The TDP-43 targets include CTNND1, MEF2D, BIM, AP2, CNTFR, MADD, CDK5RAP2, KIF2A, KIF1B, SOX9, TLE1, TNIK, UNC5C, etc., but their direct relevance to neuro degeneration remains to be elucidated.58 Second, TDP-43 shuttles between the nucleus and cytoplasm, but mostly remains in the nucleus.59 In several neurological diseases including frontotemporal dementia (FTD), ALS, and Alzheimer’s disease, TDP-43 is accumulated in cytoplasmic inclusions in the patient’s brain, which is subsequently ubiqutinated, cleaved, and abnormally phosphorylated.60,61
Third, TDP-43 plays an important role in axonal transport of mRNAs. In normal human brain, binding of TDP-43 to 3’ UTR of mRNAs is 10-fold enriched in the cytoplasm compared to the nucleus, suggesting a role of TDP-43 in mRNA stability and transport.58 TDP-43 interacts with two distinct groups of proteins constituted of a nuclear/splicing cluster and a cytoplasmic/translation cluster, suggesting that TDP-43exerts different roles in the nucleus and cytoplasm.62 One of the cytoplasmic functions of TDP-43 is to constitute cytoplasmic mRNP granules and guide delivery of target mRNA from the soma to distal axonal compartments including neuromuscular junction (NMJ).59 They also proved that TDP-43 mutations in ALS impair transport of the target mRNAs to distal axonal compartments. Pathomechanisms of TDP-43 mutations can thus be attributed to defects in splicing of target pre-mRNA due to compromised spliceosome formation, and/or defects in distal axonal transport of target mRNAs. Both abnormal nucleo cytoplasmic shutting of TDP-43 and abnormal cytoplasmic aggregates of TDP-43 potentially exert additional deleterious effects on the nuclear and cytoplasmic roles of TDP-43. Tauopathies are a characteristic pathological feature of neurodegenerative disorders, including Alzheimer’s disease, where abnormal accumulation of tau proteins occurs due to alteration in its metabolism. At least three types of alterations are observed; aberrant splicing of exon 10 of tau protein, missense mutation, and aberrant hyperphosphorylation.63,64 Glycogen synthase kinase (GSK-3) is a key enzyme that positioned at the convergence of pathways that are misregulated in Alzheimer’s disease and other tauopathies. Several modes of regulation of GSK-3 have been demonstrated including tau hyperphosphorylation, modulation of presenilin, and amyloid toxicity.63,64 In addition, GSK-3 regulates splicing of tau exon 10 by modulating SC35 phosphorylation and intracellular distribution, which is altered in some tauopathies.63
PSEN2 encoding the presenilin-2 (PS2) is one of Alzheimer’s disease (AD)-associated genes.65 Aberrant skipping of exon 5 generates a truncated deleterious isoform (PS2V), which accumulates as visible PS2V bodies at high frequency in the hippocampus of sporadic AD patients.66 High mobility group A1 a protein (HMGA1a) had been demonstrated as a splicing regulator of PS2, which binds to a specific sequence in exon 5 preceding 5’ splice site.67 Induced expression of HMGA1, observed in the brain of sporadic AD patients, result in aberrant skipping of PSEN2 exon 5, which is caused by an impaired dissociation of U1 snRNP from the 5’ splice site.67,68
Recently, mutations in RBM20 gene have been shown to be associated with human dilated cardiomyopathy (DCM).69,70 Deep sequencing of the cardiac transcriptome revealed aberrant splicing of the titin gene (TTN).71 The detected aberrant TTN transcript was previously identified to be due to a loss-of-function mutation in RBM20 gene.69,70 Subsequent analysis revealed actual cis-elements on TTN that are recognized by the splicing-suppressive RBM20.72 Transcriptome-wide RBM20-binding sites in heart were recently reported using photoactivatable ribonucleoside–enhanced crosslinking and immunoprecipitation (PAR-CLIP) followed by high-throughput sequencing of RNA.73 PAR-CLIP revealed that RBM20 regulates splicing of a network of genes with essential cardiac functions including TTN, RYR2, LMO7, RTN4, PDLIM3, CAMK2D, LDB3, etc. Validation in patients with heart failure revealed that these genes were indeed aberrantly spliced in patients with severe heart failure who had low RBM20 expression levels in the heart. RBM20 thus plays an important role in modulation of cardiac functions and its defect predisposes to cardiomyopathy and heart failure.
Nonsense-Associated Altered Splicing (NAS) and NMD-Associated Skipping of a Remote Exon (NASRE)
Generation of a premature termination codon (PTC) sometimes causes skipping of a PTC-containing exon. This phenomenon is termed as NAS and a lot of such point mutations have been reported. For example, a nonsense mutation in exon 51 of FBN1 gene, causes exon skipping which is associated with Marfan syndrome.74 NAS can be activated through different mechanisms.75 The most common one is nuclear scanning mechanism to ensure a proper translational frame. A nonsense mutation disrupting the open reading frame can direct the splicing machinery to skip the affected exon. This type of selective exon exclusion allows retention of the residual function of a protein instead of complete degradation by NMD. In case of an alternative exon, a nonsense mutation can cause an exclusive increase of exon-skipped transcripts due to the degradation of exon-included transcripts harboring a premature termination codon. In addition, a nonsense mutation can affect a local cis-element by disrupting an ESE or compromising an important element for RNA secondary structure, which subsequently promotes exon skipping. Mutation can also promote skipping of a remote exon not the PTC-bearing exon. In congenital myasthenic syndrome (CMS), we reported a 7bp deletion in CHRNE exon 7, which promotes skipping of the preceding exon 6.76 We termed this phenomenon as NASRE. Skipping of 101-nt exon 6 generates a PTC. Due to an inherent weak strength of splice sites flanking exon 6, CHRNE normally generates an exon 6-skipped transcript, which is completely degraded by NMD.76 However, the 7bp deletion in exon 7 resumes an open reading frame in the exon 6-skipped transcript, which makes the exon 6-skipped transcript immune to NMD. Remote exon skipping is also evident in other genes, such as SLC25A20,77 DBT,78 BTK,79 MLH1,80 etc. However, NASRE is likely to be underestimated because remote exons are seldom scrutinized in investigating human diseases.
Trans-Dominant splicing defects due to cis-acting repeat expansions
An unusual mode of splicing defect arises from abnormal expansion of unstable nucleotide repeats in transcribed genomic regions. Pathogenic expanded repeats can affect splicing in two ways. First, repeat expansion in the coding region can alter the function of an encoded protein, either by loss of normal functions or gain of aberrant functions. Such type of misregulations have been observed in spinocerebellar ataxias, Huntington diseases, and oculopharyngeal muscular dystrophy.81 Second, repeat expansion in a non-coding region gains an aberrant function in RNA itself. A remarkable example of this type of disease is myotonic dystrophy (DM), caused by a CTG-repeat expansion in the 3’ UTR of DMPK (DM1), or by CCTG-repeat expansion in the first intron of ZNF9 (DM2). RNA containing expanded CUG- or CCUG-repeats forms a double-stranded RNA structure and accumulate in nuclear foci, which subsequently sequesters an important splicing regulator, MBNL1. Therefore, MBNL1 is depleted from nucleoplasm, resulting in a loss of MBNL1 function.82,83 Expanded CUG- and CCUG-repeats concurrently induce the stability of another splicing regulator CUGBP1,84 although the mechanisms are not fully elucidated yet. As a consequence, numerous alterations in splicing regulations occur in different genes including chloride channel (CLCN1), bridging integrator 1 (BIN1), insulin receptor (INSR), etc. in DM tissues.81,85 Other examples of unstable repeat expansion-related diseases are discussed in details in other review articles.81,86
Misregulated splicing in cancers
Deregulated splicing events have been demonstrated in almost all kinds of cancers. Aberrant splicing affects numerous processes of cancer development and progression including cell cycle control, initiation, invasion, metastasis, apoptosis, cellular metabolism, cellular signaling, etc.87 Cancer cells exploit the plasticity of alternative splicing to generate aberrant proteins to support development and progression of cancer cells. Cancer-associated missplicing can result from alterations in multiple physiological splicing processes. The affected processes include cis-acting splicing defects in oncogenes, tumor suppressors and any other genes related to cancers; defects in splicing trans-factors including a loss of function, a gain of aberrant function, abnormal expression, subcellular mislocalization, and altered post-translational modification; and a change in cellular environment that regulates alternative splicing. Cancer-associated cis-acting splicing defects have been reported in many genes. A notable example is BRCA1, where germline mutations are predisposed to breast and ovarian cancers. An inherited ESE-disrupting mutation in exon 18 was initially reported to cause aberrant exon skipping.88 A large NUMBer of mutations affecting splicing regulatory cis-elements have been reported later in BRCA1.89‒91 Other remarkable cis-acting splicing defects associated with cancers are found in KLF6, CDH17, KIT, and LKB1, which are discussed in detail in a relevant review article.92
trans-acting splicing defects in cancers were initially characterized in family of hnRNPs and SR proteins. A noteworthy example is hnRNP A/B family consisting of hnRNPs A1, A2, and A3. Among the members, hnRNPs A1 and A2 are significantly over expressed in a variety of cancers.93‒95 Depletion of hnRNP A1 and hnRNP A2 causes apoptosis in cancer cells, but not in normal cells, indicating a clear association of these proteins in development of cancer.94 A deeply dissected splicing target of both hnRNP A1 and A2 is the PKM gene encoding pyruvate kinase. Splicing misregulation of PKM critically affects glucose metabolism in tumor cells. Mechanistic investigation demonstrated that hnRNPs A1 and A2 along with another member of hnRNP family, PTB, bind to the PKM gene in a sequence specific manner flanking exon 9 and subsequently regulate mutual splicing of exon 9 and exon 10 to generate two spliced isoforms of PKM1 and PKM2.93 Analysis of expression level in normal and glioma samples demonstrated that the ratio of PKM2/PKM1 is significantly correlated with the level of hnRNPs A1, A2, and PTB. Depletion of these trans-factors, as well as MYC, which is a modulator of their expressions, induces a switch from PKM2 to PKM1.93 The association of PTB is also reported in ovarian cancer.96 Another member hnRNP H has also been reported to be associated with cancer, which regulates splicing aberration of IG20/MADD and RON pre-mRNAs.97
Among the SR proteins, SRSF1 is the best characterized cancer-associated factor. A lot of cancer-associated missplicing events regulated by SRSF1 have been reported to date. For example, SRSF1 binds to an ESE in RON exon 12 and promotes exon skipping to generate a functionally altered truncated protein inducing cancer invasions.98 Another example is BIN1, which is a tumor suppressor gene. Induced expression of SRSF1 promotes inclusion of exon 12a, and generates an isoform lacking a tumor suppressing activity.99 In addition, over expression of SRSF1 induces generation of an isoform MNK2b from MNK2 pre-mRNA and promotes MAPK-independent phosphorylation of eIF4E, leading to oncogenic transformation.99 Another SR protein, SRSF3 is also altered in numerous cancers. SRSF3 has been reported to be over expressed in numerous human cancers including cervical, lung, breast, liver, thyroid, stomach, skin, bladder, kidney and colon cancers100 SRSF3 regulatesp53-mediated cellular senescence to suppress tumorigenesis, through alternative splicing of TP53.101 Mechanistic analysis revealed that SRSF3 binds to exon i9 of TP53 and prevents its inclusion in its pre-mRNA splicing.101 SRSF3 also regulates alternative splicing of exon 10 of PKM gene by binding to an ESE in exon 10 thereby affecting lactate production, which is critically associated with cancer.102
In addition to hnRNPs and SR proteins, other splicing regulatory proteins are also involved in cancers. For example, an RNA-binding protein, Quaking (QKI), has been identified as a key regulator of alternative splicing in lung cancer. Down-regulation of QKI expression has been demonstrated in lung cancer, which is significantly correlated with poor prognosis.103 QKI-5, a splice variant of QKI, inhibits cancer cell proliferation and prevents inappropriate activation of the Notch signaling pathway by regulating pre-mRNA splicing of a key regulatory gene NUMB.103 Investigation of underlying mechanisms demonstrated that QKI-5 recognizes two cis-elements in NUMB pre-mRNA and suppresses inclusion of exon 12 by competing with the core spliceosome component SF1. As a result, an exon 12-excluded NUMB isoform is generated, which is unable to activate the Notch signaling pathway.
Post-translational modifications of splicing factors sometimes critically affect their functions and are also documented in cancers. A striking example is hnRNP L, which regulates tumorigenesis in lung cancer by modulating pre-mRNA splicing of CASP9 encoding caspase 9.104 Alternative splicing of CASP9 generates two variants: a long variant 9a and a short variant 9b lacking four exons. A short variant 9b is deficient for apoptotic function compared to 9a. In addition 9b competitively inhibits the activation of 9a. HnRNP L is a critical splicing suppressor in non-small cell lung cancers (NSCLS), which functions by binding to an exonic splicing silencer to generate caspase 9b.104 However, the splicing modulation by hnRNP L is not evident in non-transformed cells, suggesting a potential post-translational modification of hnRNP L specific to NSCLS. Further analysis indeed revealed the phosphorylation of a serine residue (Ser55) of hnRNP L, which is critical to regulate CASP9 pre-mRNA splicing for the tumorigenic capacity of NSCLS cells.104
Web based in silico programs to analyze splicing code
With the progress of splicing researches, a lot of in Silico programs have been developed to assess alternative splicing isoforms, cis-acting splicing code, effects of mutations on splicing, predicting functional splicing regulatory motifs and their cognate partner, strength of splice sites, RNA secondary structure, etc. We have enlisted popular programs in Table 4. We also developed an algorithm in order to uncover underestimated splicing effects due to exonic mutations at the 5’ splice site.105 For this purpose, we initially constructed 31 minigenes spanning exonic splicing mutations and analyzed the splicing consequences using splicing RT-PCR. We also scrutinized 189, 249 U2-dependent 5’ splice sites of the entire human genome. As a prediction parameter, we set out a new variable called the SD-Score, which represents a common logarithm of the frequency of a specific 5’ splice site spanning 9 nucleotides (3 nucleotides at the 3’ end of an exon and 6 nucleotides at the 5’ end of an intron). Our analysis demonstrated that SD-Score can efficiently predict the splicing consequences of these minigenes. In order to improve the prediction accuracy, we further employed the information contents (Ri). In an attempt to validate our algorithm, we scrutinized 32 additional minigenes as well as 179 previously reported splicing mutations. As an estimate, the SD-Score algorithm predicted aberrant splicing in 198 of 204 sites with a sensitivity of 97.1% and normal splicing in 36 of 38 sites with a specificity of 94.7%. An extensive simulation of all possible exonic mutations at positions -3, -2 and -1 at189, 249 5’ splice sites predicted aberration of pre-mRNA splicing in 37.8%, 88.8% and 96.8%of simulated mutations, respectively, which were all higher than we expected. Although in Silico programs are playing great roles in identifying splicing cis-elements in normal and pathological conditions, no programs have 100% accuracy. Therefore, in vitro and/or in vivo investigations of individual genes, along with in Silico analysis, are indispensable for elucidation of normal and pathological splicing regulations.
Program |
URL |
Feature |
Database of alternative splicing events |
||
Alternative splicing Database176 |
http://cgsigma.cshl.org/new_alt_exon_db2/ |
A database of alternative splicing based on published reports |
ASPicDB177 |
http://www.caspur.it/ASPicDB/ |
An annotation-based database of alternative transcript and protein isoforms |
Database of mutations and other genetic variations |
||
HGMD178 |
http://www.hgmd.org/ |
A database of human gene mutations associated with inherited diseases |
ssSNP Target179 |
http://ssSNPTarget.org/ |
A database for single nucleotide polymorphisms |
dbSNP180 |
http://www.ncbi.nlm.nih.gov/SNP/ |
A comprehensive database of SNPs |
1000 genome181 |
http://browser.1000genomes.org/ |
1000 genome project |
NHLBI ESP |
http://evs.gs.washington.edu/EVS/ |
NHLBI exome sequencing project |
HGVD182 |
http://www.genome.med.kyoto-u.ac.jp/SnpDB/ |
Human genetic variation database (HGVD) |
Tools to evaluate splice sites (5’ splice site, 3’ splice site, and branch point) |
||
Human Splicing |
http://www.umd.be/HSF/ |
A tool to evaluate the splicing effect of a mutation and to identify splicing motifs in human sequences |
Finder183 |
||
SD score105 |
http://www.med.nagoya-u.ac.jp/neurogenetics/SD_Score/sd_score.html |
A tool to predict the effect of a mutation at 5’ splice site |
MaxEntScan184 |
http://genes.mit.edu/burgelab/ |
A tool to score authentic and possible splice sites in a given sequence with different models |
maxent/Xmaxentscan_scoreseq.html |
||
Analyzer Splice |
http://ibis.tau.ac.il/ssat/SpliceSiteFrame.htm |
An algorithm to calculate the scores of donor and acceptor sequences |
DBASS5187 |
http://www.dbass.org.uk/ |
A database for 5’ splice site aberration |
DBASS3187 |
http://www.dbass.org.uk/ |
A database for 3’ splice site aberration |
Splicing regulatory elements and splicing factors assessment tools |
||
Splice Aid188 |
http://www.introni.it/splicing.html |
A database of experimentally proven RNA target motifs bound by splicing proteins in humans |
SpliceAid2189 |
www.introni.it/spliceaid.html |
A database of tissue-specific human splicing factors and RNA target motifs |
http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=homeb |
A tool to identify exonic splicing enhancers |
|
A tool for identifying candidate ESEs in vertebrate exons |
||
FAS-ESS194 |
http://genes.mit.edu/fas-ess/ |
A tool to predict ESS motifs |
SFmap195 |
http://sfmap.technion.ac.il |
A tool for motif analysis and prediction of splicing factors |
RNA secondary structure assessment tools |
||
mFold196 |
http://www.bioinfo.rpi.edu/applications/mfold/rna/form1.cgi/ |
A tool to predict the secondary structure of single stranded nucleic acids |
sFold197 |
http://sfold.wadsworth.org/ |
A tool to predict RNA secondary structure, andfor the rational design of RNA-targeting nucleic acids |
pFold198 |
http://www.daimi.au.dk/~compbio/pfold/ |
RNA secondary structure prediction tool using stochastic context-free grammars |
Table 4 Databases and web-based programs for analyzing splicing mutations and alternative splicing
Analyses of abnormal splicing code in human diseases not only uncover the underlying maladies of splicing regulation in pathological conditions, but also allow us to obtain an insight in splicing controls in physiological conditions. The advances in research on splicing aberrations in numerous diseases also provide us knowledge to identify mutations that had been otherwise difficult to ascertain. In addition, understanding physiological and pathological splicing mechanisms also paves the way for development of therapeutic strategies. We have shown that the complex and intricate nature of alternative splicing events confers the transcriptome and proteome diversity and facilitates biologically beneficial processes including molecular evolution. The complex and intricate nature, however ironically makes alternative splicing highly susceptible to deleterious malfunctions causing diseases. For a particular splicing defect in a particular pathological condition, elucidation of the underlying misregulation enables us to estimate if the splicing defect is a direct cause or an indirect modifier in the process of disease development. Elucidation of the underlying mechanisms of splicing perturbations also tells us potential therapeutic targets such as RNAs, RNPs, and auxiliary associated factors. By modulating these targets, we may be able to reverse the aberrant splicing to a physiological state. Evidently, advances in molecular targeting therapy for alternative splicing are growing at a steady pace and are moving forward from the laboratory bench to the clinic.
The works performed in our laboratory were supported by Grants-in-Aid from the MEXT and MHLW of Japan.
Author declares that there is no conflict of interest.

©2015 Rahman, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.