Journal of
eISSN: 2373-4469


Research Article Volume 2 Issue 1
1Blokhin Russian Cancer Research Center, Russia
2Dana-Farber Cancer Institute, USA
Correspondence: Elena A Shestakova, Blokhin Russian Cancer Research Center, Kashirskoe shosse 24, 115478, Moscow, Russia, Tel +7 499 612 8072
Received: January 22, 2015 | Published: February 6, 2015
Citation: Shestakova EA, Nakatani Y. Characterization of histone predeposition complexes from different cellular compartments. J Investig Genomics. 2015;2(1):25-28. DOI: 10.15406/jig.2015.02.00017
The composition of cytoplasmic and nuclear histone H3.1 predeposition complexes was analyzed in this study. These complexes are used for the building of nucleosome, a basic unit of chromatin. Core histones H3.1, H4, histone chaperones and histone acetyl-transferases were identified using mass spectrometry in both nuclear and cytoplasmic complexes. Importin-beta was revealed as specific component of cytoplasmic complex and large subunit of histone chaperone CAF-1 p150 as specific component of nuclear complex.
Keywords: histone h3.1, nucleosome assembly, importin-beta, histone chaperones, mass spectrometry
RC, replication coupled; RI, replication independent; TAP, tandem affinity purification; TSA, trichostatin A
DNA replication involves the chromatin assembly on new DNA strands which involves the formation of new nucleosomes, the basic unit of chromatin. Nucleosome consists of the tetramer of H3/H4 histones and two dimers of histones H2A/H2B surrounded by DNA segment.1 Some histones composing nucleosome exist in one isoform such as histone H4, while other histones exist in several isoforms such as histone H3 (H3.1 and H3.3). Histone H3 is of special interest as different isoforms are involved in different key cellular processes: H3.1 isoform is involved in DNA replication maintaining the genome integrity and H3.3 isoform is involved in transcription which comprises the first step of gene expression. Several histone predeposition complexes were purified and characterized. Nuclear complex containing the histone chaperone CAF-1 and histone variant H3.1 that is involved in the replication-coupled (RC) histone deposition during S-phase of cell cycle and nuclear complex containing histone chaperone HIRA and histone variant H3.3 that participate in the replication-independent (RI) histone deposition during transcription were purified from HeLa cells.2,3 Cytoplasmic histone predeposition complexes were not extensively characterized as nuclear ones.
In this study histone H3.1 predeposition complexes were purified from both cytoplasm and nucleus of HeLa cells and compared using mass spectrometry analysis and immunoblotting. This analysis showed that ASF1A/B histone chaperone is a component of both cytoplasmic and nuclear complexes while large subunit of CAF1 chaperone, CAF1p150 is present only in nuclear complex. Importin-beta3 was identified as a part of cytoplasmic complex and not of nuclear one indicating that it dissociated when the complex entered the nucleus. Experiments with TSA, a histone deacetylase inhibitor, demonstrated that increased acetylation of histone H3.1 and possibly of other proteins allows the formation of H3/H4 tetramer in cytoplasm and nucleoplasm instead of dimers of H3/H4 histones identified in this and previous studies. This study provides new possibilities to develop medicaments directed against epigenetic targets and especially acetylated histones helping to treat cancer, one of the most harmful diseases of our days.
H3.1 containing complexes purification
H3.1-containing complexes were purified essentially as described in.4 Nuclear and cytoplasmic extracts were prepared from suspension growing HeLa cell line expressing H3.1 histone fused with C-terminal Flag- and HA-epitope tags (e-H3.1). Then, cytoplasmic and nuclear H3.1-containing complexes were purified from corresponding extracts by immuno precipitation on anti-Flag antibody-conjugated agarose beads, eluted with Flag peptide and were further affinity purified on anti-HA antibody-conjugated agarose beads followed by the elution with HA peptide.
Mononucleosomes purification
Nuclear pellets were homogenized and resulting oligonucleosomes were digested with micrococcal nuclease to obtain mononucleosomes as described in.5 e-H3.1 containing nucleosomes were purified from mononucleosomes by immunoprecipitation with anti-Flag antibody and eluted with Flag peptide as described in.4
Acetylation assays
Trichostatin A (TSA) was added to suspension of HeLa cell line expressing e-H3.1 at final concentration of 100 nM and at indicated time points 200 ml aliquots of cell culture were collected and mixed nuclear and cytoplasmic complexes were purified by immunoprecipitation either with anti-Flag antibody or with anti-Flag antibody followed by anti-HA antibody.
Silver staining, western blottin
Were carried out using standard techniques.
Mass spectrometry: SDS-PAGE gels with separated protein complexes were stained with Coomassie Blue 250, protein bands were cut from gels, digested with trypsin and analyzed with LC-MS/MS mass spectrometry as described in.6
Reagents: The following antibodies were used: anti-p150 antibody (H300; Santa Cruz Biotechnology); anti-ASF1 antibody (gift of Dr. Genevieve Almouzni); anti-CAF1p48 antibody (05-523, clone 3-255; Upstate Biotechnology); anti-importin beta (gift of Dr. Bauer-Mattaj); anti-histone H3 antibody (C-16; Santa Cruz Biotechnology); anti-acetyl-histone H3 antibody (Millipore, Upstate); anti-HA antibody (HA.11; Covance); anti-MCM2 antibody (BL248; Bethyl Laboratories); anti-MCM4 antibody (BL257; Bethyl Laboratories); anti-MCM6 antibody (BL262; Bethyl Laboratories); anti-MCM7 antibody (BL265G; Bethyl Laboratories). For tandem affinity purification anti-Flag agarose (A-2220; Sigma) and anti-HA agarose (sc-805AC; Santa-Cruz Biotechnology) were used. Suspension HeLa cells were cultivated in Jocklik modified MEM (M-0769; Sigma).
Purification of cytoplasmic and nuclear predeposition histone h3.1 complexes
Earlier it was demonstrated that histone H3.1 is deposited in the nucleosome as a distinct predeposition complex found in the nucleoplasm. To extend these studies we purified histone H3.1-containing complexes both from nucleoplasm and cytoplasm using as sources of these complexes nuclear and cytoplasmic extracts obtained from HeLa cells growing in suspension culture. HeLa cells that we used expressed Flag/HA-tagged histone H3.1 and were described in.3 Nuclear and cytoplasmic complexes containing histone H3.1 were purified from corresponding cell extracts using tandem affinity purification (TAP) approach as described in Materials and Methods. Briefly, corresponding extracts were first applied on agarose beads coupled with anti-Flag antibodies, Flag/HA-H3.1-containing complex was eluted with Flag peptide, then subsequently loaded on agarose beads coupled with anti-HA antibodies and eluted with HA peptide. Silver stained gel containing nuclear and cytoplasmic complexes obtained with TAP is shown on Figure 1A.
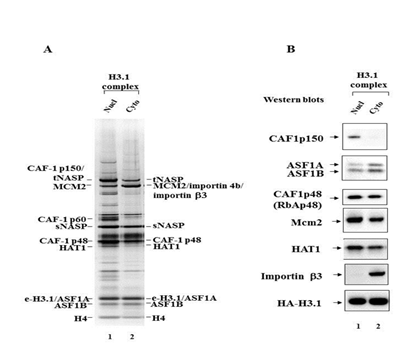
Figure 1 Mass spectrometric and immunoblotting analyses of nuclear and cytoplasmic H3.1 histone predeposition complexes.
Figure 1A Silver staining of the H3.1 nuclear (lane 1) and cytoplasmic (lane 2) predeposition complexes obtained by tandem affinity purification on anti-Flag and anti-HA antibodies coupled agarose beads from the extracts of transduced HeLa cells. The proteins identified by mass spectrometric analysis are indicated.
Figure 1B Immunoblot analysis of the H3.1 nuclear (lane 1) and cytoplasmic (lane 2) predeposition complexes. Complexes were analyzed by immunoblotting with anti-CAF1p150, anti-ASF1A/B, anti-CAF1p48 (RbAp48), anti-Mcm2, anti-HAT1, anti-importin beta3, anti-HA antibodies. The latter antibody was used to reveal the epitope-tagged histone H3.1.The patterns of nuclear and cytoplasmic complexes partially overlap although some protein bands are specific either for nuclear or cytoplasmic complexes. We applied mass spectrometric analysis in order to determine the protein composition of nuclear and cytoplasmic H3.1 complexes. This analysis revealed the following components in cytoplasmic H3.1 complex: Flag/HA-tagged histone H3.1, histone H4, histone chaperones ASF1A and ASF1B, histone acetyltransferase HAT1, small subunit of histone chaperone CAF-1 p48, MCM2 component of MCM complex involved in the initiation of DNA replication, sNASP, tNASP and importin beta3. In nuclear H3.1 complex the following components were identified: Flag/HA-tagged histone H3.1, histone H4, histone chaperones ASF1A and ASF1B, histone acetyltransferase HAT1, small and large subunits of histone chaperone CAF-1: CAF1 p48 and CAF-1 p150, MCM2 component of MCM complex involved in the initiation of DNA replication, sNASP, tNASP. Among the differences that we found between nuclear and cytoplasmic H3.1 complexes were the presence of importin beta 3 in the cytoplasmic complex and its absence in the nuclear complex and the presence of large subunit of histone chaperone CAF1 p150 in the nuclear complex and its absence in the cytoplasmic complex (Figure 1A). The protein composition of nuclear and cytoplasmic complexes revealed with mass spectrometry was confirmed with Western blotting technique (Figure 1B). Taken together these findings shed light on the mechanisms of successive formation of H3.1 core complex in the cytoplasm, its transport into the nucleus through the nuclear pores with the participation of importin beta 3, dissociation of the importin in the nucleus on the inner side of the nuclear membrane and acquisition of the large subunit of histone chaperone CAF1 p150 with the following formation of the mature nuclear histone H3.1 complex which will be directly deposited in the chromatin forming the nucleosome
MCM protein complex involved in the initiation of DNA replication is found in both cytoplasmic and nuclear h3.1 histone predeposition complexes
Given that using mass spectrometry we identified MCM2 component of MCM protein complex known to be involved in the initiation of DNA replication in both cytoplasmic and nuclear H3.1 histone predeposition complexes we further used Western blotting technique to analyze the presence of other components of MCM complex in both histone H3.1 predeposition complexes. Using antibodies developed against MCM2, MCM4, MCM6 and MCM7 components of MCM protein complex we confirmed the presence of MCM2 in cytoplasmic and nuclear H3.1 histone predeposition complexes and demonstrated the presence of almost all subunits of MCM complex in both H3.1 histone predeposition complexes and the absence of MCM complex subunits in cytoplasmic and nuclear mock complexes (Figure 2). These findings clearly indicate that H3.1 predeposition complex or at least its sub fraction is involved in the initiation of DNA replication and probably its progression.

Figure 2 MCM proteins in cytoplasmic and nuclear H3.1 histone predeposition complexes.
Cytoplasmic (lane 4) and nuclear (lane 6) H3.1 histone predeposition complexes were obtained by tandem affinity purification on anti-Flag and anti-HA antibodies coupled agarose beads from the extracts of transduced HeLa cells. Cytoplasmic mock (lane 5) and nuclear mock (lane 7) purifications were made from the corresponding extracts of non-transduced HeLa cells. Cytoplasmic and nuclear H3.1 histone predeposition complexes, nuclear and cytoplasmic mocks were analyzed together with different amounts of cytoplasmic (lanes 1, 2, 3) and nuclear (lanes 8, 9, 10) extracts from transduced HeLa cells by immunoblotting with anti-MCM2, MCM4, MCM6 and MCM7 antibodies.
H3 and H4 histones exist as dimers in both nuclear and cytoplasmic h3.1 complexes
It has been shown in previous studies2,3 that H3 and H4 histones exist as dimers in nuclear H3.1 complexes. We extended these studies to the cytoplasmic H3.1 complex and demonstrated that H3 and H4 histones exist as dimers in cytoplasmic H3.1 complex as in the nuclear one (Figure 3).
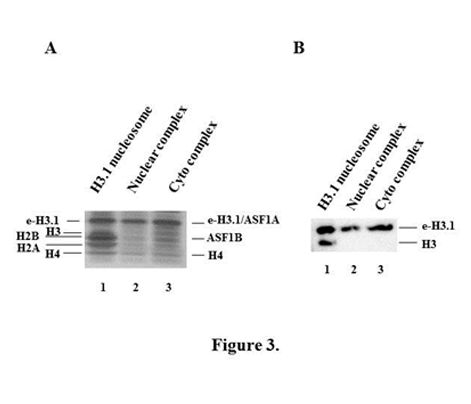
Figure 3 H3 and H4 histones exist as dimers in both nuclear and cytoplasmic H3.1 complexes.
Figure 3A Silver staining of SDS-PAGE with e-H3.1-containing mononucleosomes (lane 1) and e-H3.1-containing nuclear (lane 2) and cytoplasmic (lane 3) predeposition complexes. e-H3.1-containing mononucleosomes were purified from nuclear pellets of transduced HeLa cells and e-H3.1–containing nuclear and cytoplasmic complexes were obtained from either nuclear or cytoplasmic extracts of these cells by tandem affinity purification on anti-Flag and anti-HA antibodies coupled agarose beads. The positions of epitope-tagged histone H3.1 (e-H3.1), endogenous histone H3.1, histones H2B, H2A and H4 are indicated for the mononucleosome and the positions of epitope-tagged histone H3.1 (e-H3.1), histone H4 and ASF1A/ASF1B are indicated for the nuclear and cytoplasmic histone predeposition complexes.
Figure 3B: Immunoblotting of e-H3.1-containing mononucleosomes and e-H3.1-containing histone predeposition complexes. e-H3.1-containing mononucleosomes (lane 1) and nuclear (lane 2) and cytoplasmic (lane 3) histone H3.1 predeposition complexes were analyzed by immunoblotting with anti-histone H3 antibody.
Histone deacetylases prevent histone h3/h4 tetramer formation in cytoplasm and nucleoplasm
To analyze the dependency of tetramer formation on the acetylation status of histone H3 we applied a specific inhibitor of histone deacetylases, Trichostatin A (TSA), to the culture of HeLa cells for different time intervals, prepared cell extracts at different time points after TSA treatment and purified from a series of such extracts the mixtures of nuclear and cytoplasmic histone H3.1 predeposition complexes. Surprisingly, we observed the appearance histone H3/H4 tetramer in the presence of the inhibitor of histone deacetylases, TSA indicating that histone deacetylases prevent histone H3/H4 tetramer formation in cytoplasm and nucleoplasm (Figure 4).
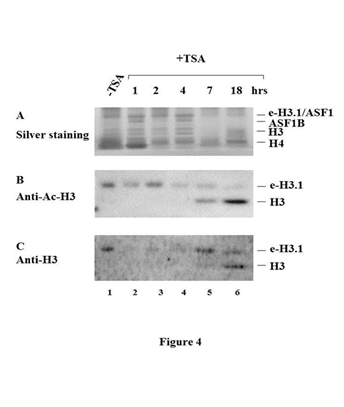
Figure 4 Histone deacetylases prevent histone H3/H4 tetramer formation in cytoplasm and nucleoplasm.
The mixtures of nuclear and cytoplasmic histone e-H3.1 predeposition complexes were purified by immunoprecipitation with anti-Flag antibody (B, C) or anti-Flag antibody followed by anti-HA antibody (A) from the extracts of transduced HeLa cells prepared at different time points after TSA treatment. Nuclear/cytoplasmic histone e-H3.1 predeposition complexes purified from untreated cells (lane 1), from cells after 1 hour (lane 2), 2 hrs (lane 3), 4 hrs (lane 4), 7 hrs (lane 5), 18 hrs (lane 6) of TSA treatment were separated by SDS-PAGE and either stained with silver (A) or analyzed by western blots with anti-Ac-H3 (B) and anti-H3 antibodies (C).
In present study using mass spectrometry and Western blotting we compared the compositions of cytoplasmic and nuclear H3.1 histone predeposition complexes purified from transduced HeLa cells expressing Flag/HA-tagged histone H3.1. This analysis demonstrated similar composition of histone H3.1 predeposition complexes present in two main compartments of the cell, cytoplasm and nucleus. The major differences that we observed in these complexes are the following: we identified the importin beta 3 in cytoplasmic complex which was not found in nuclear one and we demonstrated the presence of the large subunit of histone chaperone CAF-1 p150 only in the nuclear complex. These findings allow us to propose the model according to which the core complex is formed in the cytoplasm that contains the dimer of histones H3/H4, the histone chaperone ASF1A/B, the histone acetyltransferase HAT1, the small subunit of histone chaperone CAF-1 p48, MCM complex involved in the replication of DNA and importin beta 3. With the help of the importin beta 3 the core complex is transported into the nucleus through the nuclear pore, loses the importin beta 3 on the inner side of the nuclear pore and acquires the additional subunits of histone chaperone CAF-1 (p150 and p60) forming the complete histone H3.1 predeposition complex (Figure 5) that assembles the nucleosome on the replicating DNA.
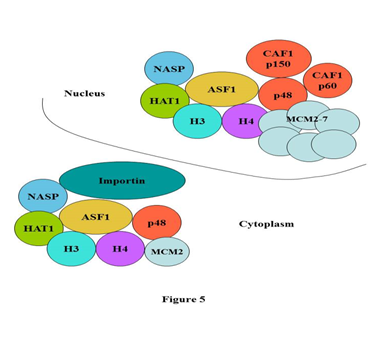
Figure 5 Model of histone H3.1 predeposition complex formation from cytoplasm to nucleus.
According to this model the core complex is formed in the cytoplasm that contains the dimer of histones H3/H4, the histone chaperone ASF1A/B, the histone acetyltransferase HAT1, the small subunit of histone chaperone CAF-1 p48, MCM complex involved in the replication of DNA and importin beta 3. With the help of the importin beta 3 the core complex is transported into the nucleus through the nuclear pore, loses the importin beta 3 on the inner side of the nuclear pore and acquires the additional subunits of histone chaperone CAF-1 (p150 and p60) forming the complete histone H3.1 predeposition complex that assembles the nucleosome on the replicating DNA.
Taken together, this study opens up vast possibilities to develop new medicaments directed against epigenetic targets and especially acetylated histones. This direction of research is developing rapidly in our days. One of the best examples of such investigations is the development of very specific inhibitors ((+)-JQ1) of BET bromodomain-containing chromatin adaptor proteins subfamily.7 Such inhibitors abrogate the MYC gene transcription resulting in significant anti-tumor activity in xenograft models of Burkitt’s lymphoma and acute myeloid leukemia.8 Therefore, these inhibitors have perspective clinical utility as MYC oncogene is a very powerful inducer of different malignancies including Burkitt’s lymphoma and acute myeloid leukemia. Moreover, these studies point the new directions and roads in the development of medicaments targeting the epigenetic mechanisms of cancer initiation and progression therefore improving targeted anti-cancer therapy. Intensive and extensive introduction of such medicaments in clinics would be one of the promising ways in the eradication of cancer, one of the most harmful diseases of our days.
This research was supported with grants from NIH (GM065939-02) and Human Frontier Science Program to Dr. Yoshihiro Nakatani (Dana-Farber Cancer Institute and Harvard Medical School, Boston, USA). We are grateful to Dr. Steven Gygi and Dr. Ross Tamaino in the Taplin Biological Mass Spectrometry Facility (Harvard Medical School, Boston, USA) for mass spectrometry analysis.
Author declares that there is no conflict of interest.

©2015 Shestakova, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.