Journal of
eISSN: 2373-4469


Here we decipher the logic underlying a key functional motif that defines the landscape of Alu repetitive elements. Alu positions 7-10 lie within the A Box, a site necessary for its transcription by Pol III and subsequent retrotransposition. We show that this site originated as a mother motif (CGCG) that gave rise to four daughter and ultimately four granddaughter motifs. The sequences of the progeny are all dictated by methylation-driven cytosine deamination of the mother. These finding provide the mechanistic basis for diversification and expansion of Alu, a major event in primate evolution.
Alu repeats are short interspersed elements1‒4 that are of great interest because they arose along with primates ~65 million years ago, because they are restricted to the primate lineage, and because they are the most abundant mobile elements in primates.5‒8 Consequently, Alus are a potential "reason" that primates differ from other species. Alus continue to expand throughout the human genome, even today as an Alu is inserted at a new position in the human genome every twenty or so births. The 1.1 million Alus in the genome have an immense influence on human biology.5 Insertion of Alu elements cause numerous human genetic diseases ranging from cancer to familial hypercholesterolemia.5 Over the last decade it has also become clear that Alus can contain tens of thousands of functional transcription factor binding sites.9 For this reason, Alus are believed to have played a role in establishing larger gene regulatory networks.9‒11 The role of Alu also extends to the RNA world since bioactive Alu elements are transcribed to free monomeric Alu-RNAs (small cytoplasmic: scAlu)5 and because dozens of microRNAs are transcribed through Alu-dependent RNA polymerase III (Pol III) transcription.12
The mechanistic logic underlying the expansion of the lineage of Alu remains unclear. It was originally hypothesized that Alu elements arose from a single master gene,13 but this notion has evolved to the current belief that a small subset of Alus remain competent for retrotransposition, and therefore serve as source genes for further propagation.5 Thus, Alu elements that were at one time, or still are, active are considered master or "source" genes.5,14 It is not possible to define source Alus with the well known JSY hierarchical classification scheme. This classification system is based on 20-30 diagnostic positions that undergo neutral mutation,15‒17 which allows for stratification of Alus into families of different age, the J (oldest), S (intermediate) and Y (youngest), and makes it possible to understand the molecular archeology of Alu. Almost by definition, however, the JSY classification scheme is independent of the changes to functional sites brought about by evolutionary pressure. Hypermutable CG doublets are a profound example of this because they are excluded from the sequence analyses used for JSY classification. The C of ‘CG’ is a preferred site of methylation, an epigenetic process linked to chromosome remodeling and gene silencing. Indeed, Alus contain nearly one-third of the genome’s CpG sites and they play a major role in epigenetic silencing.18‒22
Here we consider the possibility that methylation-driven cytosine deamination is a guiding event in the propagation and functional diversification of Alu. Specifically we focus on positions 7-10 of Alu (labeled according to homology with the sequence of the 7SL gene). These positions lie within the Alu A box, which functions along with a downstream B box to create the canonical binding site for Pol III.23 In the human 7SL gene, a presumed ancestor of Alu,6,24 positions 7-10 are CGCG, a motif that can be viewed as two adjacent and hypermutable CpG sites. By the process of cytosine methylation, and subsequent deamination, these sites could be converted to TpG (or CpA on the antisense strand).18,22 Some of the first evidence for this possibility came from a study by Zemojtel et al.,9 who showed that this particular CGCG in Alu is deaminated to create a vast number of functional binding sites for p53. Thus, full development of the methylation-driven deamination landscape of a "mother" CGCG sequence, shows that it would be converted to a select set of only four daughter motifs (one deamination) and four additional granddaughter motifs (two deaminations). One the other hand, random mutation of a four nucleotide stretch would theoretically yield a normal distribution of 256 possible sequences. To distinguish between these two extremes, we tallied the motifs at position 7-10 in a set of 8,422 representative dimeric human Alu sequences extracted from the human genome (Supplemental Figure 1). While 1,844 Alus contained motifs that are outside of the deamination landscape, the vast majority of the Alus (6,538) contained either the mother CGCG, or one of the eight daughter or granddaughter motifs that arise by deamination (Figure 1A) (Figure 1B). Essentially same distribution was observed in the entire human genome when all 1,091,321 Alus were analyzed. This result leads to the inescapable conclusion that CpG deamination has been the major determinant in the sequence at Alu positions 7-10. It is almost inconceivable that such a constrained distribution could have arisen in any other manner.
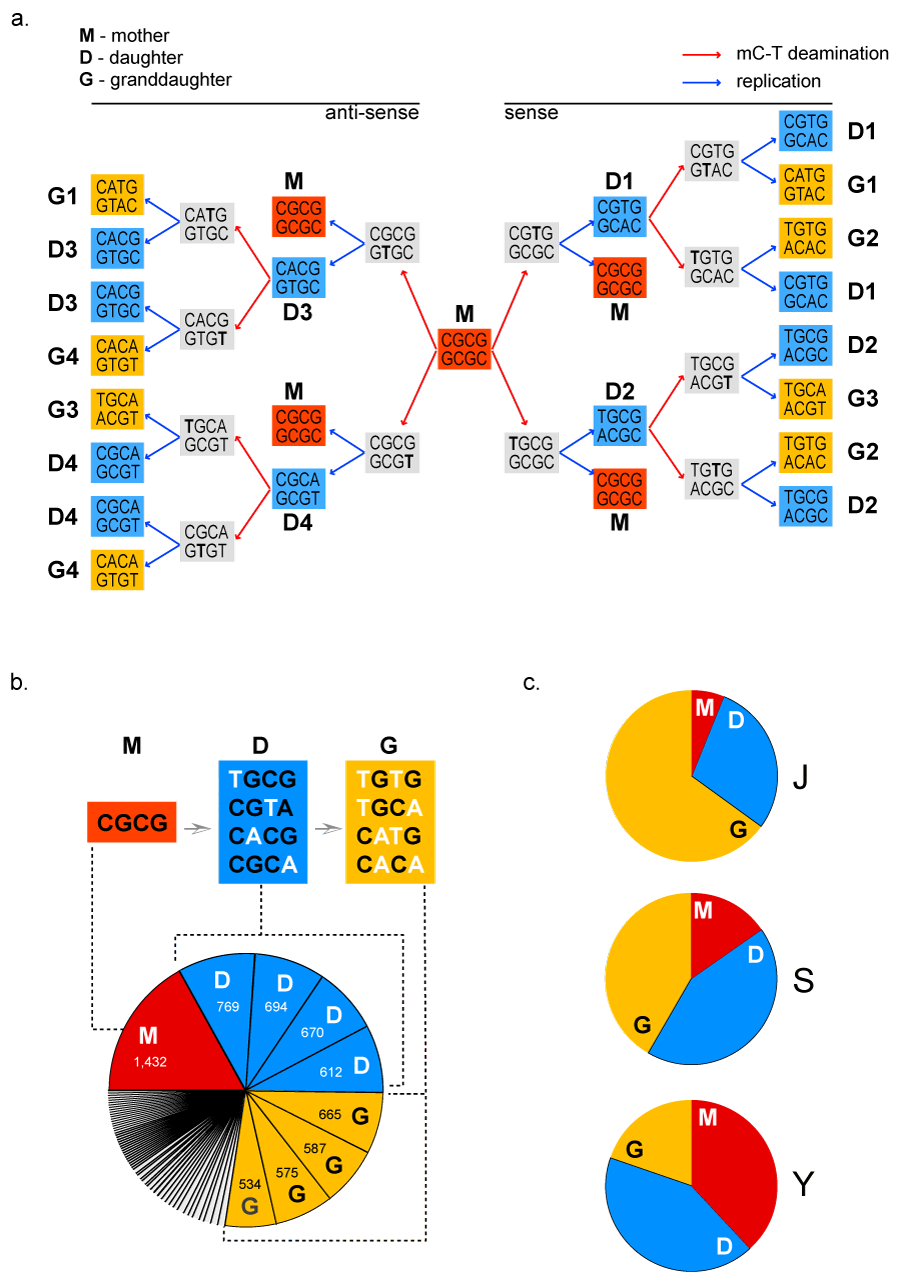
Figure 1 Methylation-driven Deamination Defines the Logic of Alu.
Figure 1a The methylation-driven diversification of positions 7-10 in Alu begins with a mother CGCG motif (M-red), which is methylated and then deaminated at one position (red arrows leading to grey boxes). Replication of each deaminated motif leads to daughter motifs (D, blue boxes). Daughter motifs are methylation and deaminated (red arrows), and then replicate (blue arrows) to create granddaughter motifs (G, gold boxes). The motifs derived from the sense strand of CGCG are shown on the right, and those from the anti-sense strand on the left.
Figure 1b The distribution of each mother (red) daughter (blue) and granddaughter (gold) motif within the 6,538 dimeric Alus was tallied (confirmed also in the context of whole genome when 1,091,321 were analyzed). Motifs that fall outside of the deamination scheme are uncolored in the pie chart. The sequence and site of deamination for each motif represented in the pie chart is indicated in boxes above.
Figure 1c The fraction of each type of motif that is classified as a J, S or Y Alu was determined with the CENSOR tool (http://www.girinst.org/censor/) (see Supplemental Figure 2 and details).
If methylation-driven deamination shaped the sequence at positions 7-10 in the Alu population, one would also anticipate a time-dependent conversion of the mother into the daughter, and ultimately the granddaughter motifs. To look for such a trend, we examined the distribution of motifs in the J (oldest), S (intermediate) and Y (youngest) families of Alu.5,17 The 6,538 test Alus were segregated into J, S and Y families with the online CENSOR tool, and the fraction of mother, daughter and granddaughter motifs in each Alu family was calculated. In the Alu J family, the mother CGCG motif represents only a small fraction of the elements (6%). A substantial number of the J Alus have been converted to daughter motifs (29%), and an even higher fraction (65%) into granddaughter motifs. In the Alu S family, which originated somewhere between 65 and 35million years ago, 15% of the 7-10 motifs are mother, 43% daughter and 42% granddaughter motifs. In the Y family, which arose only 2million years ago,5,16 the mother CGCG motif is 38%, the daughter motifs represent 42%, but granddaughter motifs are only 20% (Figure 1C). Thus, we observe an age-dependent increase in mother elements, and a concomitant decrease in granddaughter motifs. This trend can certainly be taken as strong evidence that the mother CGCG has a higher rate of retrotransposition than the progeny. If either of the progeny were more active, one would expect to observe that the frequency of progeny would be higher than that of the mother in younger Alus as well as their overall sequence conservation (e.g. fossil-Alu-monomers with CGCG-motif should be mostly preserved in primates; Figure S3). While more complicated scenarios could conceivably explain the observed distributions, the simplest interpretation of the data is that methylation-driven deamination of positions 7-10 shaped the Alu landscape during primate evolution.
There is a strong line of reasoning to suggest that the mother CGCG motif is predominant in source Alus. Since Pol III transcription of Alu is necessary for retrotransposition, any active Alu must support interaction with Pol III. A key site for this interaction is the A box, which encompasses positions 7-10 in Alu.23 Interestingly, work in the early 1990s showed the granddaughter motifs CATG and TGCA are unable to support Pol III transcription,19 so these progeny are unlikely to be active. In our search of the literature, we found only one daughter motif (CGTG) reported to support Pol III transcription.25 Along similar lines, the 40 human Alu elements with demonstrated retrotransposition activity in cells, all contain the mother CGCG motif.26 While no comprehensive study has been conducted on the efficiency by which all daughter and granddaughter motifs can support Pol III transcription, and/or retrotransposition, the available biochemical data are entirely consistent with the idea that Alus with the mother CGCG are most active, and that the progeny, particular granddaughters, are less active. The fact that most Alus of the Y family, which are presumed to be the most active, contain the CGCG motif is also consistent with the idea that Alus with the mother motif are optimal "source" Alus.
Based on the observations put forth here, we propose a new three-dimensional projection of the phylogeny of Alu that takes into account both the methylation-driven logic described here, and the canonical JSY classification scheme (Figure 2). This projection essentially illustrates how extant Alus containing mother, daughter and granddaughter motifs have arisen, and how they project onto a flat landscape representing the JSY classifications. This cone-like projection begins to map the underlying biochemical mechanisms subject to evolutionary pressure that shaped the Alu landscape. Because Alu elements are so prevalent in primate genomes, the methylation-driven logic uncovered here is likely to have been a major driving force in primate evolution.
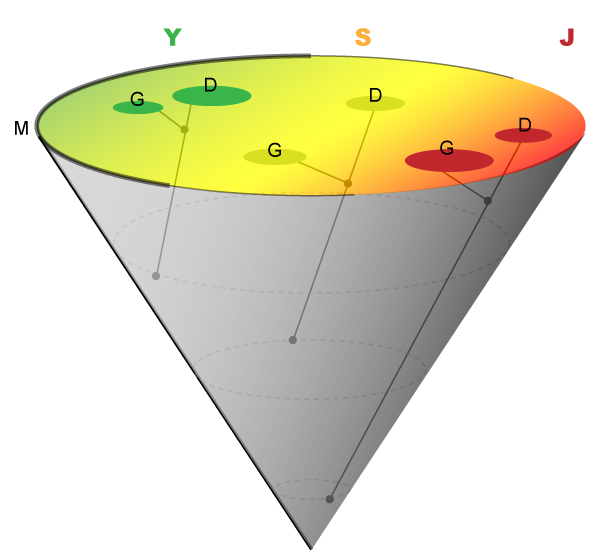
Figure 2 Intersection of the Alu 7-10 Lineage and the JSY Landscape.
The Alu 7-10 lineage can be visualized as a cone projecting onto a surface representing the JSY landscape (base of the cone). The J, S, and Y families are represented as a continuum of colors (J-red; S-yellow; Y-green). Alus containing the mother motif (M) originate at the apex of the cone and project upward as the entire outer surface of the cone. The relative distribution of the mother is represented by the thickness of the base of the cone. Alus containing daughter (D) and granddaughter (G) motifs project away from this surface just as they would in a two-dimensional phylogenetic tree. Darker spots indicate positions where daughter and granddaughter Alus project onto the JSY landscape.
To obtain a representative set of Alus we focused on the regions upstream of human genes. We created a database containing the untranslated regions (UTRs) extending from the translation start site (ATG) to -5,000 in 17,532 human genes. These sequences were extracted using the genome browser at UCSC (http://genome.ucsc.edu/cgi-bin/hgGateway, 2006 assembly - hg18,)S1 and were then deposited in a Structured Query Language (SQL)-database. Each UTR in the database was matched to all coding sequences from the RefSeq database (http://www.ncbi.nlm.nih.gov/RefSeq). This allowed us to define first ATG-codon of each coding sequence and then to create positional coordinates for each UTR. A database containing this information was compiled and it contains ~92 megabases (~3% of the human non-coding genome).
Alu sequences were computationally extracted from UTRs in this database using "collocation" searches like those used in statistical language processing to classify segments of text based on the presence of two or more separate words.S2 By way of analogy then, we sought to identify "Alu paragraphs" by searching for "words" common to all Alus. Three sequence motifs were used as the search words; the sequence that encodes the SRPS3,S4 binding site in Alu RNA, and the motifs encoding the A- and B-boxes.S5,S6 The A-box is positioned seven nucleotides upstream of the SRP binding motif and the B-box is positioned 46 nucleotides downstream yielding an Alu signature of "|A-box|−|SRP|−|B-box|" (Supplemental Figure 1, top panel).
All sequences corresponding to the SRP binding site were extracted from the database of UTRs. Then, within this set, we identified all sequences that contained an A- and B-box at a fixed distance from one another (as indicated in the top panel). This procedure extracted 12,440 Alus, which included both dimeric (~300 nucleotides) and monomeric Alu sequences (~150 nucleotides) (Supplemental Figure 1, Venn diagram). Since Alu monomers might include aberrant or truncated sequences, we elected to exclude them from the analysis. This was accomplished by a second collocation search for the poly-A tail that exists between the monomers of full-length Alu’s. As a result of this filtering, we obtained dimeric 8,422 Alu sequences that were used for this study (Supplemental Figure 1, Venn diagram, blue). Alu-7–10-tailing was also performed on all human Alus {hg19: 1,091,321}. It showed essentially the same distribution. The majority of the Alus (710,598: 65.11%) contained the CGCG motif or one of the eight potential methylation driven derivatives of this motif. The frequency of CGCG, which we call the mother (M), was found in less than 10% of all Alus, whereas the majority contained a motif derived from deamination at one site, which we call daughters (D), or at both sites, which we call granddaughters (G). Furthermore, of the 35% of Alus that lack one of the eight noted motifs, nearly half of these (45%) contain a motif that can be derived by a single point mutation in one of the G motifs. Consequently, the majority of the sequence landscape at 7-10 in Alu has been shaped by methylation driven cytosine deamination (manuscript in preparation).
The online classification of Alu elements was performed by a program available on a public server ("CENSOR", www.girinst.org/censor) dedicated to analysis of repetitive elements as deposited in RepBaseS7 and originally classified by RepeatMasker (another well-known tool for library-based repeat identification by A. F. Smit, R. Hubley and P. Green; http://www.repeatmasker.org). RepBase Update, the most comprehensive database of repetitive element consensus sequences, is also available for use in the CENSOR or BLASTN search step. RepBase Update is compiled and maintained by the Genetic Information Research Institute.S8,S9 Both programs (RepeatMasker and CENSOR) perform similarity searches based on local alignment using precompiled libraries of consensus or representative sequences of repeat families in RepBase. CENSOR, like RepeatMasker, is designed to locate and mask regions in genomic sequences that correspond to known repetitive elements.S10 CENSOR uses the fast and sensitive similarity search program WU-BLAST (W. Gish; http://blast.wustl.edu). Optionally, the BLASTN or BLASTX programs of the WU-BLAST package can be used directly instead of CENSOR. The authors are grateful to Jerzy Jurka and the GIRI for providing the RepBase Update database and the CENSOR program. The CENSOR program is being developed and maintained by Oleksiy Kohany at GIRI. The authors also thank the TIGR Plant Repeat Database Team for making their databases freely available. The CENSOR server permits screening repeats in DNA sequences from all eukaryotic species represented in the database by comparing them to the most recent version of RepBase Update and returning output back to the user via the browser (if used in a web/remote host-mode, see a snapshot of the web-CENSOR-input interface) or after download of CENSOR program on a local PC/CPU (access of the compressed *tar.gz file requires user registration). We submitted as input the sets of Alu-sequences as grouped in the M/D/G ‘families’ all compiled in FASTA-format and uploaded to CENSOR in a batch mode (i.e. uploaded from a local file). Detailed information about CENSOR input/output is presented in "Help/Information" main menu at (www.girinst.org/censor). CENSOR can be run in three different sensitivity modes. The WU-BLAST parameter settings corresponding to these modes are listed in the online Tutorial. Certain parameters (word size, E-vAlue threshold, gap penalties) of the direct WU-BLAST searches can also be adjusted by the user (see the online Tutorial for details). We used the default options of CENSOR. After the web-based CENSOR returned its output for each of the M/D/G Alu-submissions, the tabular annotations were extracted and converted into MS-Excel files, where Alus in each M/D/G group were segregated into J, S and Y families as reported by CENSOR.
A simplified branching order of the major Alu subfamilies is shown. Branching points correspond to the average age of individual JSY subfamilies in million of years (Myr), all presumably originating from 7SL RNA.24 The results of the present study suggest branching of the Alu lineages based on CpG mutations at position 7-10 (Figure 2). These mutations in a Mother (M) Alu give rise to Daughter (D) and Granddaughter (D) Alus. Because deamination at CpG is much faster than non-CpG mutations, an alignment of the two evolutionary processes results in MDG branching for each of the JSY lineage. Daughters that mutate at single position are labeled ‘Sm’. Fossil-Alu-Monomers (FAMs) with the mother (M) motif (M-FAM), the human tRNA-Ala, human 7SL RNA gene (hs7SL) and the BC200 RNA gene (hsBC200) were aligned with CLUSTAL W (1.83) and their phylogenetic relationship by Phylogeny.fr (www.phylogeny.fr) (manuscript in preparation). The orthologs of the seven M-FAMs were tracked in nine primates. Absent orthologs at the same chromosomal location (broken synteny) is shown by the absence of a gray square in the matrix. The percent of the indels for each M-FAM are shown with bar chars on the right (insertions: light gray and deletions: dark gray) (manuscript in preparation).
None.
Author declares that there is no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.