Journal of
eISSN: 2373-6453


Research Article Volume 2 Issue 7
1Abbott Molecular, Des Plaines, USA
2University of Texas Medical Branch, USA
3BioMONTR?Labs, Research Triangle Park, USA
Correspondence: Danijela Lucic, Abbott Molecular, Des Plaines, 1350 E Touhy Ave, Des Plaines, IL 60018, USA, Tel (224) 361 7124, Fax (224) 361 7646
Received: November 30, 2015 | Published: December 8, 2015
Citation: Lucic D, McClernon A, Fangling Xu, Liang X, Weaver S, Dong J, Cloherty GA, McClernon D (2015) Workflow Optimization of the ViroSeqTM Human Immunodeficiency Virus-1 (HIV-1) Genotyping System. J Hum Virol Retrovirol 2(7): 00071. DOI: 10.15406/jhvrv.2015.02.00071
Background: Current therapy guidelines recommend resistance testing to guide therapy in antiretroviral (ARV) treatment naive patients as well as patients with suboptimal virologic responses or virologic failure.
Objective: This study evaluated workflow optimization and automation improvements to the ViroSeq HIV-1 Genotyping System through the use of the semi-automated extraction platforms (Abbott Molecular Inc. m2000sp and bioMérieux Inc. NucliSens easyMAG) for sample extraction, NanoDrop 2000 Spectrophotometer (Thermo Scientific, CA) and Agilent Bioanalyzer 2100 (Agilent Technologies, Germany) for amplicon quantitation and Applied Biosystems Instruments (ABI) 3500 (Life Technologies, Ca) for increased sequencing throughput (Table 1).
Methods: Clinical plasma samples were processed in parallel with Viroseq HIV-1 Genotyping System per manufacturer instructions along with above described automated options. Resistance associated mutations from the different approaches were evaluated and assessed for clinical impact.
Results: All samples tested in this study gave valid results regardless of the procedure used. Samples extracted with m2000sp demonstrated resistance- associated mutations in 24/30 samples (80%) while complete drug resistance concordance was observed in 29/30 samples (97%). Ninety one percent (10/11) of the samples extracted with easyMAG had 98.7% concordance at the nucleotide level and 100% concordance for drug resistance mutations. No apparent difference was observed in sequence quality between manual and semi-automated extractions and comparable drug resistance profiles were obtained regardless of the extraction method.
Conclusion: The ViroSeq HIV-1 Genotyping System is compatible with the semi-automated extraction systems, alternative amplicon quantitation devices as well as with high throughput sequencing platform such as the ABI 3500 sequencer. This study also indicated that the assay can accurately sequence HIV-1 virus below 1,000 copies/mL. Significant workflow and turnaround time advantages were observed in this study with the automation of the Viroseq HIV-1 Genotyping System.
Keywords:ViroSeq HIV-1, Genotyping system, NanoDrop 2000 spectrophotometer, Bioanalyzer 2100, ABI 3500 instrument, m2000sp, CD4 T cells
HIV-1, Human Immunodeficiency Virus; ARV, Antiretroviral; ABI, Applied Biosystems Instruments; HAART, Highly Active Antiretroviral Therapy; NRTI, Nucleoside Reverse Transcriptase Inhibitors; NNRT, Non-Nucleoside Reverse Transcriptase Inhibitors; PI, Protease Inhibitors; PR, Protease; RT, Reverse Transcriptase; RAM, Resistance Associated Mutations
There were approximately 35.3 [32.2–38.8] million people living with HIV-1 in 2012.1 ART therapy has converted HIV-1 infection from an almost universally fatal illness to a chronic, manageable disease.2 However, since latent infection of CD4 T cells acts as a reservoir, enabling lifelong persistence and re-emergence of the virus, the eradication of HIV-1 has not been possible to date.3,4 Therefore; the primary goals for initiating ART are to:
Effective treatment of HIV-infected individuals with ART is highly effective at preventing transmission to sexual partners.6 In 2012 more than 9.7 million people living with HIV-1 were receiving an ART in low- and middle-income countries.7 However, less than one-third of HIV-infected individuals in the United States have suppressed viral loads.8 If antiretroviral viral drug levels are suboptimal, the risk of developing ARV resistance is high due to the high rate of HIV-1 replication and the lack of proofreading capacity in the transcriptase enzyme.2
Transmitted drug resistance can seriously limit future therapeutic options. North American and European treatment guidelines recommend resistance testing for all treatment-naive patients prior to the initiation of highly active antiretroviral therapy (HAART).9 Genotypic testing is recommended as the preferred resistance testing to guide therapy in ARV-naive patients or to guide therapy in patients with suboptimal virologic responses or virologic failure while on first or second regimens.10 This provides the opportunity to avoid ineffective drug combinations and allows for individualized optimization of HAART.9
Standard genotypic resistance assays provide information on resistance to nucleoside reverse transcriptase inhibitors (NRTIs), non-nucleoside reverse transcriptase inhibitors (NNRTIs), and protease inhibitors (PIs) by identifying resistance-related mutations in the viral genome. One such test is the ViroSeq™ FDA HIV-1 Genotyping system11 (Celera Diagnostics, US) which is capable of sequencing the whole protease (PR) and partial reverse transcriptase (RT) region (up to codon 335) of the polymerase gene. The assay covers all the well-defined protease inhibitor and RT inhibitor resistance-related mutations. Current resistance methodologies require significant technical effort and assay turnaround time is between 2-3 days depending on sample extraction, throughput and sequencer used by the laboratory.
The ViroSeq workflow consists of manual extraction, reverse transcription and polymerase chain reaction, amplicon purification and quantitation, cycle sequencing and sequence purification followed by data collection and analysis. As the number of people gaining access to ART continues to increase dramatically the need to greater automation of the current commercially available genotypic tests is becoming more important.
This study evaluated workflow optimization and automation improvements to the ViroSeq HIV-1 Genotyping System through the use of the semi-automated extraction platforms (Abbott Molecular m2000sp and bioMérieux NucliSens easyMag) for sample extraction, NanoDrop 2000 (Thermo Scientific, CA) and Agilent Bioanalyzer 2100 (Agilent Technologies, Germany) for amplicon quantitation and Life Technologies ABI 3500 instrument for increased sequencing throughput (Table 1).
Step |
FDA Approved Kit Method |
Automated Optimization Options |
NA Extraction |
Ethanol Precipitation |
m2000sp or easyMag (Automated Extraction Systems) |
Reverse Transcription |
Kit Method Amplification |
Kit Method Amplification |
PCR |
Kit Method Amplification |
Kit Method Amplification |
PCR Purification |
Enzymatic Method |
QIAquick PCR Purification (Spin Columns) |
Post PCR QC/Quantification |
Agarose Gel (Visual Estimation Using Mass Ladder and Brightness) |
NanoDrop 2000 (Spectrophotometer) and/or Agilent 2100 Bioanalyzer (Visual Estimation Using Mass Ladder) |
Normalization/Dilution |
Dilution based on Estimation of Concentration in Set Volumes |
Normalization to ~6ng/ul Concentration based on Nanodrop Concentration or Dilution based on Estimation of Concentration in Set Volumes |
Cycle Sequencing |
Kit Method PCR |
Kit Method PCR |
Cycle Sequencing Purification |
Ethanol/Sodium Acetate Precipitation, Isopropanol Clean-Up, or Centri-Sep 96 Well Plates |
Ethanol/Sodium Acetate Precipitation, Isopropanol Clean-Up, or Centri-Sep 96 Well Plates |
Sequence Detection |
ABI 3100, ABI 3130xl |
ABI 3100, ABI 3130xl, ABI 3500 |
Table 1 Schematic of Workflow Optimization for the ViroSeq HIV-1 Genotyping System Compared to the FDA Approved Kit Method
Residual samples collected for routine clinical monitoring were used during the study evaluation and as such no additional ethics review was required. For all samples, the manufacturer’s instructions were followed at each step unless otherwise specified. Nucleic acid extraction with the m2000sp was evaluated (n=30) with viral loads ranging between 1,000-400,000 copies/mL and the easyMAG was evaluated (n=13) with viral loads ranging between 2,000-1,000,000 copies/mL as well as a subset of samples (n = 6) that were diluted to 300 copies/mL and 500 copies/mL and compared to the original undiluted sample (30,000 copies/mL). All nucleic acid extractions carried out on the m2000sp were performed using a modified Total Nucleic Acid protocol provided by the manufacturer (reference: TDNA-VS Plasma-LL-500-070- v02272012). All nucleic acid extractions carried out on the NucliSens easyMAG were performed using off-board lysis and the easyMAG Generic 2.0.1 protocol. Extraction efficiencies between the manual extraction and semi-automated extractions were also evaluated by running the purified products on a standard 1% agarose gel (data not shown).
Amplicon quantitation was evaluated either by standard 1% agarose gel, NanoDrop 2000 Spectrophotometer or Agilent Bioanalyzer 2100. Agarose gel amplicon quantitation was performed per manufacturer’s instructions. The alternate amplicon quantitation methods required RT-PCR product to be purified using the QIAquick PCR purification kit and manufacturer’s instructions (Cat No 28104 and 28106, Qiagen, Maryland). One microliter of purified amplicon was analyzed spectrophotometrically using the NanoDrop 2000 and subsequently each sample aliquot was normalized to 6 ng/uL with molecular grade water. This diluted sample was then utilized for set up of the cycle sequencing reactions. Agilent amplicon analysis was performed using one microliter of purified amplicon and the Agilent DNA 7500 Kit and manufacturer’s instructions (Cat No 5067-1506, Agilent Technologies and Germany).
Direct sequencing and data analysis were performed using either the Viroseq FDA approved IVD configuration (ABI 3100 and 3130XL) or ABI 3500 instrument. For the ABI 3500 analysis data collection software v1.0, a 50 centimeter array and POP7 were used with a standard sequencing run module. Sequence analysis software v5.4, base caller KB v1.4.1.8 and mobility file KB_3500_pop7_BDTv1.mob were required for data analysis. Viroseq HIV-1 Genotyping software v2.8 (IVD) was used to generate resistance reports for all resistance analysis (Figure 1).
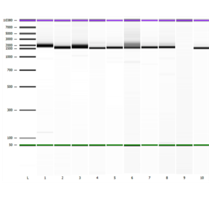
Figure 1 EasyMAG extraction Lanes 1-9 represent amplicon banding pattern seen on Agilent 2100 System using the DNA 7500 kit. Lane 10 represents the ViroSeq Kit Positive Control. Band Sizes: Lane 1 (1,972 bp), Lane 2 (1,666 bp), Lane 3 (1,694 bp), Lane 4 (1,577 bp), Lane 5 (1,643 bp), Lane 6 (1,651 bp), Lane 7 (1,692 bp), Lane 8 (1,717 bp), Lane 9 (negative sample, no amplification) and Lane 10 (1,569 bp).
Resistance associated mutations (RAM) were found in 24 samples extracted with manual/m2000sp comparison (80%) while 6 samples demonstrated no RAM across methods (data not shown). Complete drug resistance concordance was observed in 29/30 samples (97%). One sample with resistance-associated mutations yielded a mixture of L74I/V (A/T/G) when nucleic acids were extracted manually and L74V (T/G) when nucleic acids samples were extracted by the automated m2000sp instrument. Of the 13 samples extracted with easyMAG, one sample did not produce amplified product and one sample did not generate a report due to lack of the double stranded coverage; as such they were excluded from the analysis. For 10/11 samples (91%) extracted with easyMAG there was 98.7% concordance at the nucleotide level and 100% concordance for drug resistance mutations. For 1/11 (9%), a V75I mutation was identified by the reference method (based on unidirectional coverage) while no mutations were identified for the easyMAG extracted sample (data not shown). Samples diluted to 300 copies/mL and 500 copies/mL and extracted with easyMAG had >99% concordance at the nucleotide level when compared to the original undiluted sample ≅30,000 copies/mL (data not shown). PCR products analyzed on the Agilent 2100 Bioanalyzer were the correct size and density at ~1.8kb (Figure 2).
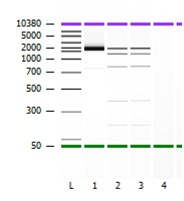
Figure 2 Representation of Agilent 2100 Bioanlayzer image. Lane L represents Agilent Ladder. Lane 1 represents purified PCR amplicon (reported @ ~1.8 kb and 13.9ng/ul). Lane 2 and Lane 3 represent ViroSeq mass ladder (Top band = ~2.0 bp, Second band = ~1.2 bp, Third bank 0.8 kb.
Differences in nucleic acid yield were observed between the manual and semi-automated methods (data not shown) however this did not lead to any apparent difference in sequence quality between methodologies. Further, comparable drug resistance profiles were obtained regardless of the extraction method.
Significant workflow advantages were observed in this study with the use of the semi-automated extraction methods and NanoDrop 2000 platforms. Sample extraction for ViroSeq™ HIV-1 Genotyping system per current manufacturer instructions takes approximately 3 hours for 12 samples with half of that time being accounted as technician hands on time. Utilizing a semi-automated instrument for nucleic acid extraction reduced the hands on time to less than 30min when processing up to 24 samples.
The results of this study imply that significant improvements in workflow are possible with the use of the semi-automated platforms in conjunction with the ViroSeq™ HIV-1 Genotyping system, with no impact to results reported. Standard manual sample extraction takes approximately 3 hours for 12 samples with half of that time being accounted as technician hands on time. Utilizing a semi-automated extraction platform for ViroSeq HIV-1 sample extraction improves technician efficiency by allowing technicians to perform other laboratory tasks while extraction is ongoing and reduces the hands-on time from 1.5 hours to less than 30 min. The number of samples extracted can also be scaled up without significantly increasing technologist hands-on time. Automated sample extraction also reduces manual manipulation of samples and reagents, which can introduce pipetting errors that can impact reproducibility as well as the risk associated with repetitive stress injury.
The differences in nucleic acid yield observed may be due to extraction efficiencies as the manual method requires concentration of the sample into a pellet prior to nucleic acid extraction while the semi-automated methods do not incorporate any upfront concentration steps. These extraction differences would suggest that gel amplicon quantitation may not be an optimal method for amplicon quantitation and devices such as the NanoDrop 2000 should be considered. In addition, NanoDrop removes the subjective band evaluation and provides fast and accurate quantitation down to 1-2 ng/uL, compared to gel electrophoresis which can only detect 5-10 ng/uL. Laboratories which are looking to further optimize the current ViroSeq HIV-1 workflow and eliminate the use of ethidium bromide can substitute agarose gel with Agilent 2100 Bioanalyzer which provide both amplicon size and concentration information. Alternatively, the ViroSeq mass ladder may be run concurrently allowing the end user to perform amplicon dilution based on band intensity (see example provided in Figure 2). The gel image may still be appropriate for troubleshooting purposes especially in cases where laboratory amplicon degradation or contamination is suspected.
The results of this study demonstrate that the ViroSeq HIV-1 Genotyping System is compatible with the semi-automated extraction systems, NanoDrop 2000, Agilent Bioanalyzer and the ABI 3500 sequencer. These alternative configurations offer considerable workflow advantages over the FDA approved configuration of manual extraction / agarose gel/ABI 3100/3130X platforms. While additional studies may be required to confirm the impact of detecting nucleotide mixtures at lower viral concentrations, these findings indicate that the assay can accurately sequence HIV-1 virus below 1,000 copies/mL.
Funding and reagents for these studies was provided by Abbott Molecular (Des Plaines, IL), bioMérieux Inc. (Durham, NC) and bioMONTR Labs (Research Triangle Park, C).
DL and GC are employees and shareholders of Abbott Inc., AM, DM and JD has received funding from Abbott Inc., FX, XL and SW has nothing to disclose.
AM, FX, XL carried out the laboratory studies, participated in the sequence alignment and drafted the Manuscript. DL and GC conceived of the study. DL, DM, GC, SW, JD participated in the design of the study and drafted the manuscript. All authors read and approved the final manuscript.
None.

©2015 Lucic, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.