Journal of
eISSN: 2373-6453


Review Article Volume 1 Issue 2
Department of Microbiology and Immunology and Center for Molecular Virology and Translational Neuroscience, Institute for Molecular Medicine and Infectious Disease, Drexel University College of Medicine, USA
Correspondence: Brian Wigdahl, Department of Microbiology and Immunology, Institute for Molecular Medicine and Infectious Disease, Drexel University College of Medicine, 245 N. 15th Street, MS #1013A, Philadelphia, PA 19102, USA, Tel 215-762-7598, Fax 215-762-1955
Received: May 06, 2014 | Published: June 2, 2014
Citation: Dahiya S, Liu Y, Williams J, Pirrone V, Nonnemacher MR, et al. (2014) Role of Downstream Elements in Transcriptional Regulation of the HIV-1 Promoter. J Hum Virol Retrovirol 1(2): 00006. DOI: 10.15406/jhvrv.2014.01.00006
The human immunodeficiency virus type1 (HIV-1) promoter, the long terminal repeat (LTR), is central to regulating many aspects of viral life cycle dynamics. After viral integration into the host genome, the interactions of host and viral factors with the regulatory elements in the LTR govern viral gene expression, contributing to formation of either a productive transcriptional state or an inactive one. The molecular architecture of the LTR, especially due to the nucleosomal packaging, presents DNA binding elements to cellular transcription factors not only to sites that are upstream of the transcriptional start site but also to regions of the promoter that are downstream of the start site. The importance of the downstream sites has been appreciated specifically in imposing a fine-tuning on the tightly regulated process of gene expression under the control of HIV-1 LTR. This report provides a comprehensive review of the HIV-1 LTR interactions with the transcription factors in the downstream region and summarizes the functional impact of these events on viral gene expression.
Keywords: HIV-1, LTR, Transcription factor binding sites
HIV-1, Human Immunodeficiency Virus Type 1; LTR, Long Terminal Repeat; C/EBP, CCAAT/Enhancer Binding Protein; ATF/CREB, Activating Transcription Factor/Cyclic AMP Response Element Binding; TFBSs, Transcription Factor Binding Sites; NFAT, Nuclear Factor For Activated T Cells; DSE, Downstream Sequence Element; DBF, Downstream Binding Factor; GLS, Gag Leader Sequence; EMSA, Electrophoretic Mobility Shift Assays; TAR, Trans Activation Response; TPA/PMA-responsive element, Tetra decanoyl phorbol 13-Acetate Responder Element or Phorbol Ester-Responsive Element; VPA, Valproic Acid; SAHA, Suberoyl Anilide Hydroxamic Acid; CNAC, Comprehensive Neuro AIDS Core Center
Human immunodeficiency virus type 1 (HIV-1) RNA and protein expression relies heavily on interaction of host cellular transcription factors and viral factors with cis-acting regulatory elements within the HIV-1 promoter or long terminal repeat (LTR) (reviewed in.1-3). The HIV-1 LTR is divided into three regions: unique 3' (U3), repeat (R), and unique 5' (U5) (Figure 1). Many cis-acting regulatory elements located upstream of the transcriptional start site (designated+1) have been identified as modulators of HIV-1 proviral gene expression. These elements contain the core promoter region: three Sp1 sites and TATA box; the enhancer region, which includes two nuclear factor-κB sites; and the modulatory region, which includes three CCAAT/enhancer binding protein (C/EBP) sites, the activating transcription factor/cyclic AMP response element binding (ATF/CREB) region, and upstream stimulatory factor (reviewed in.4-6).
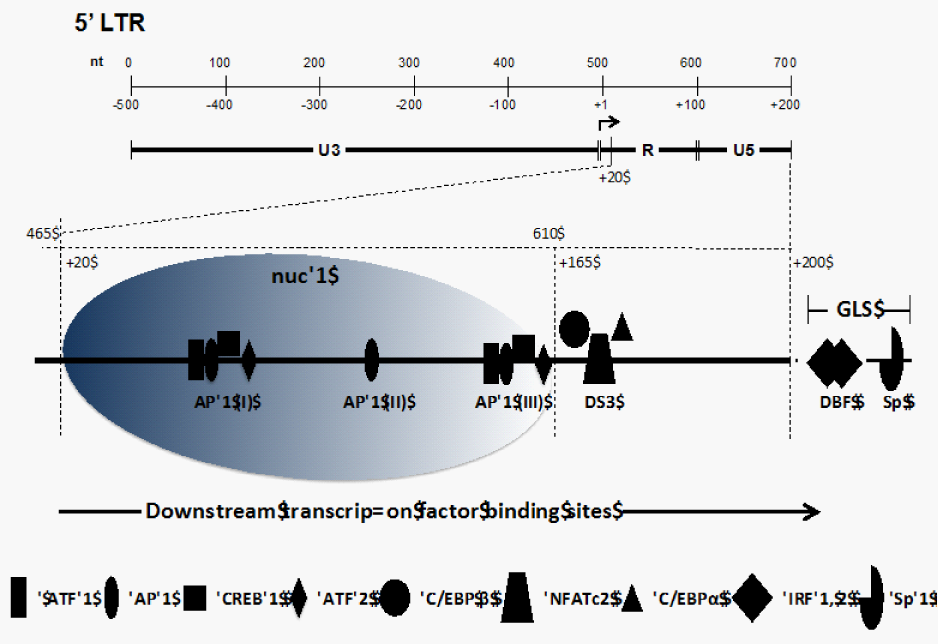
Figure 1 Organization of the 5' LTR of the HIV-1 genome. The top section shows the nucleotide positioning within the LTR and positioning relative to the transcriptional start site (+1). The LTR is divided between the U3, R, and U5 regions. The bottom section shows the nucleosomal packaging in the region downstream of the start site along with the cellular transcription factors binding to the R and U5 regions of the LTR.
Like the transcription factor binding sites (TFBSs) located upstream of the transcriptional start site, TFBSs present downstream have been shown to act in the process of transcriptional regulation. Such sequences have been reported in both TATA-containing and TATA-less promoters from viral promoters to Drosophila core promoters.7-11 The HIV-1 LTR contains several important downstream regulatory TFBSs including AP-1 motifs, an AP-3-like (AP-3L) motif, C/EBP/NFAT (nuclear factor for activated T cells) downstream binding site (DS3), two downstream sequence element (DSE) sites, one downstream binding factor (DBF-1), and two Sp1 sites in the U5 and gag leader sequence (GLS) region.5,12-17 It is also important to note that the nucleotide sequences of some of the aforementioned sites, including AP-3L, DBF-1, Sp1, and C/EBP/NFAT, are well conserved in different HIV-1 isolates, even across different subtypes, which indicates that these sites are important in HIV-1 transcription and replication.12,14 Such downstream sequences that influence gene expression in HIV have also been shown to be well conserved in some plant viruses including rice tungro bacilliform virus,18 where they modulate the level of RNA pol II processivity and HSV-1 where they are required for late viral gene expression.19 This review summarizes the well-established downstream TFBSs in the HIV-1 LTR and also describes their functional importance in the highly regulated process of HIV-1 transcription (Table 1). Most studies that have been cited herein utilized electrophoretic mobility shift assays (EMSA) and in vitro foot printing to establish binding site phenotypes and transient expression systems to define the functional role of the downstream TFBSs. This review does not discuss one of the important functional regions, the trans activation response (TAR) element found within the R (nt+1 to +60) region because this element has been extensively reviewed recently.5,20
Binding Site |
Position |
Binding Protein(s) |
Binding Sequence |
Function |
Reference |
AP-1 (I) |
+87 to +94 |
AP-1 (fos/jun), possibly ATF/CREB proteins |
TTGAGTGC |
Positive regulator of basal transcription; enhancer of replication |
|
AP-1 (II) |
+118 to +125 |
AP-1 (fos/jun), possibly ATF/CREB proteins |
TTGTGTTGA |
Positive regulator of basal transcription; enhancer of replication |
|
AP-1 (III) |
+155 to +163 |
AP-1 (fos/jun), possibly ATF/CREB proteins |
TTTAGTCAG |
Positive regulator of basal transcription |
|
DS3 |
+158 to +175 |
NFATc2, C/EBP family members |
TCAGTGTGGAAAATC |
Positive regulator of transcription |
|
AP-3L |
+162 to +177 |
NFAT |
AGTGTGGAAAATCTCT |
Positive regulator of basal transcription |
|
DSE-1 |
+88 to +98 |
Complex consisting of ATF-1, ATF-2, CREB (which does not bind the element directly), fos, and jun |
TGAGTGCTTCA |
Mediates responses to activation signals |
|
DSE-2 |
+156 to +164 |
Complex consisting of ATF-1, ATF-2, CREB (which does not bind the element directly), fos, and jun |
TTAGTCAGT |
Mediates responses to activation signals |
|
DBF-1 |
+200 to +219 |
IRF-1, IRF-2 |
CTTGAAAGCAAAGGGAAAC |
Positive regulator of basal transcription |
|
Sp |
+270 to +278 |
Sp1 |
GAGGCGAGG |
Necessary for replication; possible chromatin remodeling (nucleosome 2 positioning) |
|
Sp |
+281 to +289 |
Sp1 |
GAGGCGACT |
Necessary for replication; possible chromatin remodeling (nucleosome 2 positioning) |
Table 1 Downstream regulatory elements and their function in the HIV-1 LTR
Nucleosomal packaging within the HIV-1 LTR
After integration of the HIV-1 genome into the host DNA, two nucleosomes are assembled on the viral LTR; these have been designated nuc-0 and nuc-1. The relative positioning of these nucleosomes is fairly precise, with nuc-0 occupying nucleotide positions 40 to 200 and nuc-1 occupying positions 465 to 610 on the HIV-1 LTR 21,22 (Figure 1). Nuc-1, due to its proximity to the transcriptional start site (+1), plays an important role in the process of HIV-1 transcription, initiation, and elongation.14,21,22 It is also well established that subsequent to transcriptional activation of the integrated LTR, a large region (~255 nucleotides) downstream of the start site is free of nucleosomes due to remodeling of nuc-1.23,24 Nuc-1 remodeling is implicated as a prerequisite step in HIV-1 transcript formation. Studies have shown that phorbol esters (PMA) are able to activate HIV-1 gene expression by inducing disruptive chromatin changes in the downstream R/U5 region to allow increased binding of AP-1 and NFAT (AP-3L).14 Moreover, it is possible that some of the TFBSs that lie at the 3' boundary of nuc-1 play an important role in the positioning of nuc-1 within the downstream region of the LTR.22 Thus, the regulation of HIV-1 transcript formation is a tightly regulated process whereby the processivity of RNA Pol II may be influenced by the binding events occurring downstream of the transcriptional start site.
Activator protein-1 (AP-1) site
In addition to the three upstream AP-1 binding sites,25-27 three downstream AP-1 binding motifs have been identified in the R/U5 region of the HIV-1 LTR (Figure 1) using foot printing assays with recombinant AP-1 protein14 (Table 1). The AP-1 transcription factors belong to the basic leucine zipper transcription factor family28 and are composed of Jun/Fos heterodimers or Jun/Jun homodimers and bind to the 12-0-tetradecanoylphorbol 13-acetate responder element or phorbol ester-responsive element (TPA/PMA-responsive element).29-31 The first AP-1 motif, AP-1 (I), located at the 3' end of the R region, spans the sequence +87 TTGAGTGC +94. The second and third motifs, AP-1 (II) and AP-1 (III), respectively, are localized in the U5 region and encompass the sequences +118 TTGTGTGA +125 and +155 TTTAGTCAG +163.14 These AP-1 TFBSs differ from the consensus AP-1 sequence (TGAGTCA) by two nucleotides.28,32 Moreover, it was established that the affinities of these downstream AP-1 TFBSs for recombinant AP-1 are in the following order: AP-1 (III) > AP-1 (II) > AP-1 (I).23 C-Jun can heterodimerize with ATF/CREB32-34 or other transcription factors and bind to AP-1 sites, further increasing the diversity of transcription factors having a potential to bind these AP-1 sites.23 The functional impact of these downstream AP-1 sites has been studied and it was demonstrated that a virus containing a double mutation of the AP-1 (III) and AP-3L sites (see below) results in a 49% decrease in basal transcription compared with parental; however, these mutations exhibited a replication phenotype similar to parental virus. Conversely, mutation of all three AP-1 binding sites and the AP-3L site led to a 72% decrease in parental HIV-1 RNA production, and the replication level was also lower using peripheral blood mononuclear cells and T-lymphocyte cell lines including Jurkat T cells and SupT1 cells. These observations indicated that AP-1 (I) and AP-1 (II) sites were functionally more relevant in affecting HIV-1 transcription and replication than AP-1 (III).23
Downstream site 3 (DS3) or NFAT-C/EBP site
A recently published report12 analyzed the downstream transcription factor binding region in the LTR and found that the region from +158 to +172 (designated DS3) was conserved in 67% of unique HIV-1 subtype B LTR sequences. Moreover, the DS3 TFBS was well represented across different HIV-1 subtypes, in terms of conservation of both nucleotide sequence and nucleotide positioning within the LTR. In terms of binding phenotype analysis, using chromatin immunoprecipitation and electrophoretic mobility shift (EMS) assays on chronically HIV-1-infected cells35 established that NFATc2 and C/EBP α and β transcription factors bound DS3 in a competitive fashion, with NFATc2 demonstrating a higher relative affinity compared with C/EBP isoforms.12 Functionally, the DS3 TFBS was shown to have a positive effect on the LTR-directed transcription specifically in cells of the monocyte-macrophage lineage; no such effect was observed when a T-cell line was utilized.
AP-3-like sites
Foot printing assays using nuclear extracts from B lymphocytes have also identified an AP-3L motif, +162 AGTGTGGAAAATCTCT +177, which partially overlaps with the DS3 downstream TFBS (+158 to +172) discussed above and was shown to bind NFAT1 using nuclear extracts from A3.01 cells treated with TPA-ionomycin.14,23 Another study demonstrated the presence of NFAT in this downstream region; however, the nucleotide position of the site was not mapped specifically within the HIV-1 LTR.17 This particular region of the LTR has attracted attention because it lies at the 3' end of nuc-1 in the LTR and, as discussed previously, this nucleosome is specifically disrupted during active LTR-directed transcription. The crucial positioning of this element along with the interplay between transcription factor interactions means this region occupies a functionally relevant position within the downstream region of the HIV-1 LTR.
DSE sites
Two DSEs referred to as TPA/PMA-responsive element (TRE)/ cAMP-responsive element (CRE) were identified within the HIV-1 downstream LTR U5 region and designated DSE-1 (+88 TGAGTGCTTCA +98) and DSE-2 (+156 TTAGTCAGT +164). DSE-1 overlaps with the AP-1 motif and DSE-2 overlaps with the AP-3L site.15,16 The two DSEs exhibited sequence similarity to classical TRE and CRE sequences.16 EMS assays using nuclear extract from colonic epithelial cells showed that the DSEs formed complexes that co-migrated with DNA complexes formed with consensus TRE and CRE sites, suggesting that the DSE sites contained both TRE and CRE sites. Furthermore, super shift EMS assays showed that ATF-1, ATF-2, and CREB as well as c-Fos and JunD were all components of the DSE-1 and-2 DNA-protein complexes.15,16 However, purified ATF/CREB proteins were unable to bind to the DSE-1 and-2 sites whereas c-Jun homodimers were able to bind to DSE sites, indicating that ATF/CREB proteins may have formed heterodimers with AP-1 proteins first and then interacted with HIV-1 DSE in vivo.36
Functional studies have indicated that DSE-1 and DSE-2 are able to affect HIV-1 transcription through cellular signaling pathways involving protein kinase A (PKA) activation.23 Studies using cholera toxin, which is a potent activator of cAMP/PKA signaling,37 demonstrated that cholera toxin-mediated induction of HIV-1 LTR-directed activation is mediated by increasing CREB DNA binding to DSE-2.15,16,23,38 The ability of the DSE to bind both Jun/Fos and ATF/CREB proteins may enable these sites to integrate PKA and PKC (protein kinase C) activation signals transmitted to the viral LTR, resulting in synergistic activation of HIV-1 transcription upon cellular activation.15,36
DBF-1 sites
The downstream binding factor 1 (DBF-1) site was identified from in vitro foot printing assays using nuclear extract from B lymphocytes.14 The DBF-1 motif (+200 CTTGAAAGCAAAGGGAAAC +219) lies adjacent to the primer binding site, which is part of the untranslated leader sequence.39 The DNA sequence of DBF-1 was homologous to the interferon (IFN)-stimulated regulatory element (ISRE), which is present in the promoter of IFN-stimulated genes.14,40,41 Interestingly, experiments replacing DBF-1 with ISRE showed a reduction in both viral transcription and replication.42 Competitive EMS assays showed that the DBF complex shift was completely abrogated by both ISRE and DBF homologous oligonucleotides. Super shift assays showed that interferon-regulatory factors 1 and 2 (IRF-1 and-2) were the components of the DBF–protein complex.23,42,43
Functional assays using transient transfection showed that DBF-1 sites contribute significantly to the HIV-1 LTR basal activity in lymphocytes and HeLa cells.14 Assays using provirus containing point mutations in the DBF-1 site, which abrogated DBF-1 binding, showed a decrease in HIV-1 transcription levels, with no effect on HIV-1 replication.23 Only IRF-1 and IRF-2, from among a family of nine members,44 have been shown to bind the HIV-1 LTR, with IRF-1 being implicated in increasing the LTR-directed transcriptional activity and HIV-1 replication. This induction was shown to occur early in infection, before Tat expression.45 Additionally, IRF-1 expression can be induced by interleukin (IL)-1, IFN-γ, IL-6, and tumor necrosis factor-α (TNF-α) and can contribute to nucleosomal remodeling through recruitment of p300/CBP. Thus, the DBF site could modulate the responsiveness of HIV-1 to extracellular stimuli, including cytokines.23
Sp1 sites
Along with the well-characterized Sp TFBSs in the core region of the upstream LTR, foot printing assays using purified Sp1 protein have identified two juxtaposed Sp1 binding motifs, +270 GAGGCGAGG +278 and +281 GAGGCGACT +289, in the downstream region of the LTR. The first Sp1 binding site sequence more closely matches the high-affinity consensus Sp1 binding site sequence (5' GGGGCGGGG) than the second site. This region has several interesting features. First, this area (+270 to +290) turns out to be very conserved within different HIV isolates (a maximum 4-nucleotide difference within this region).14 Second, the Sp1 sites are located in a constitutively DNAse hypersensitive area (HS4) that also includes DBF-1. This positioning suggests that the Sp1 elements maybe involved in chromosome remodeling (namely, the positioning of nucleosome 2).
Sp1 protein binding in vivo was confirmed with studies using chronically HIV-1-infected ACH-2 and U1 cell lines. These studies also indicated that the element is constitutively bound to Sp1. Point mutations, which disrupt binding, did not significantly affect transcription, although double mutation of DBF-1/Sp1 reduced activity more than DBF-1 mutation alone.14 Provirus containing mutated Sp1 binding was unable to produce virus, suggesting this element is critical for viral replication. Interestingly, however, transient transfection using this same Sp1 mutation had little effect on transcription.23 Thus, the Sp1 elements may be involved in nucleosome positioning and viral replication and may act synergistically with other TFBSs to help positively regulate transcription.
The observations reported in this review demonstrate that transcriptional regulation via the downstream TFBSs is crucial in HIV-1 transcription, viral replication, and infectious virus production. These TFBSs introduce additional regulatory elements that warrant further study to fully elucidate the complex process of LTR-directed transcription. The molecular mechanisms by which these downstream TFBSs contribute to transcriptional regulation include recruitment of histone acetyl transferases (HAT) and ATP-dependent chromatin remodelers such as SWI/SNF (switch/sucrose non fermenting) family. NFAT family members illustrate an example of this mechanism; NFATs cooperate with AP-1 and C/EBP, which act by recruiting HATs and SWI/SNF complex.46-51 Similarly, C/EBP family members are known to interact with chromatin remodeling machinery resulting in synergistic LTR activation, for example, C/EBP β associates with CBP/p300, PCAF, SWI/SNF remodeling factors, and other co activators52-54 Another important aspect of HIV-1 pathogenesis with respect to the downstream region is the convergence of signaling pathways at this region in response to activation signals (Figure 2). The binding of transcription factors downstream of the transcriptional start site could cause additional cellular specificity1,12,23 that differentially regulates the virus in different physiological locations including the periphery and the brain. One specific example is the DS3 element that was shown to be functionally important only in cells of the monocyte-macrophage lineage with no effect yet identified in a T-cell line.12 Thus, the downstream binding sites not only increase the strength of the promoter-enhancer unit in the LTR but also provide a mechanism to broaden the viral response to extracellular stimuli and regulate transcription in a cell-type dependent manner. Moreover, the continuous evolution of HIV-155 presents a variety of nucleotide configurations of TFBSs that can respond to cellular activation signals in unique ways.
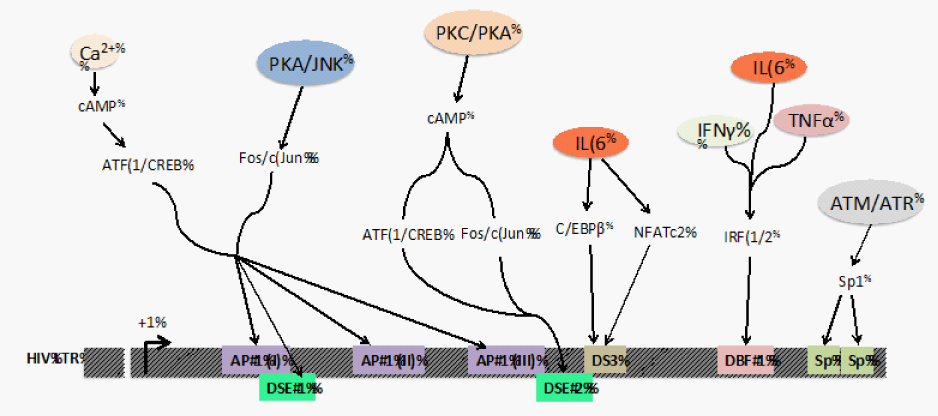
Figure 2 Integration of cellular activation signals on the downstream TFBSs in the HIV-1 promoter. The interactions at the downstream TFBSs in the LTR are dependent on cellular signals received by cytokines (IL-6, IFNγ, TNFα), activation signals from upstream kinases (PKC, PKA, JNK, ATM and ATR), and changes that regulate intracellular Ca2+ levels. These upstream signals result in post-translational modifications like phosphorylation and glycosylation on various transcription factors that trigger their nuclear translocation and exposure of their DNA binding domains. These events culminate in TF-DNA interactions that eventually recruit factors that regulate the processivity of RNA pol II on the HIV-1 LTR.
The proximity of the downstream TFBSs to the transcriptional start site also allows a direct influence on the formation of the basal transcriptional complex along with controlling RNA polymerase processivity.56-58 Thus, studying the downstream region, especially in an integrated scenario to focus on DNAse I hypersensitive areas, can provide important cues that can extend our knowledge with respect to transcriptional activation in both active and latent proviral genome pools.59The attempts to purge the latent HIV-1 reservoir have included strategies that utilize cytokines like IL-6, IL-2, and TNFα to induce a global cellular activation;60-63 HDAC1 inhibitors like valproic acid (VPA) and suberoyl anilide hydroxamic acid (SAHA) to induce transcription64,65 all of which would have impact on the downstream region of the LTR; and inhibition of DNA methylation using 5-aza-2’deoxycytidine (aza-CdR).66 Newer approaches have shown an interest in a novel group of transcriptional regulators that control viral gene expression. These include the bromodomain (BET family) inhibitors that can induce the activation of latent HIV-1 in primary cell models of latency.67-69 Thus, understanding the molecular underpinnings of transcriptional regulation would enhance the progress towards development of a cure for HIV infection.
These studies were funded in part by the Public Health Service, National Institutes of Health, through grants from the National Institute of Neurological Disorders and Stroke, NS32092 and NS46263, the National Institute of Drug Abuse, DA19807 (Brian Wigdahl, Principal Investigator), National Institute of Mental Health Comprehensive Neuro AIDS Core Center (CNAC), P30 MH-092177 (Kamel Khalili, PI; Brian Wigdahl, PI of the Drexel subcontract), and under the Ruth L. Kirschstein National Research Service Award 5T32MH079785 (Jay Rappaport, PI; Brian Wigdahl, PI of the Drexel subcontract). The contents of the paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
None.

©2014 Dahiya, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.