Journal of
eISSN: 2373-6453


Research Article Volume 9 Issue 1
MD, PhD, Institute of Progressive Medicine, South Pasadena, USA
Correspondence: W John Martin, MD, PhD, Institute of Progressive Medicine, South Pasadena CA 91030, USA, Tel 01-626-616- 2868
Received: June 19, 2022 | Published: June 24, 2022
Citation: Martin WJ. Renegade cellular genetic sequences in stealth adapted viruses: introducing a new era of virology. J Hum Virol Retrovirology. 2022;9(1):24-29. DOI: 10.15406/jhvrv.2022.09.00242
DNA sequence data have previously been obtained on an African green monkey simian cytomegalovirus (SCMV)-derived stealth adapted virus. The virus was repeatedly cultured from a patient with the chronic fatigue syndrome (CFS). The data reveal not only genetic sequences that are derived from regions of the SCMV genome, but also the unexpected presence of genetic sequences that have originated from portions of the human cellular genome. The SCMV-derived stealth adapted virus has also acquired foreign genetic sequences of bacterial origin. The focus of this article is on the potential mechanism as well as the major biological and clinical ramifications of the primate to human and subsequent human to human viral transmission of genetically unstable renegade cellular genetic sequences. Insight into this topic has come from further analysis of rhesus monkey-derived cellular sequences in the stealth adapted viruses cultured from two other CFS patients and a mixture of both rhesus and human genome-derived cellular sequences in the virus cultured from another CFS patient. The virus acquired monkey cellular sequences are subject to ongoing mutations and can be replaced by human cellular sequences, probably by homologous recombination. There is a genetic basis for many human diseases, including cancers. The potential acquisition of pathogenic cellular sequences by stealth adapted viruses may, therefore, result in some of these genetic diseases becoming infectious. Stealth adapted viruses have been cultured from patients with a range of neurological and psychiatric illnesses, yet their existence is still not officially acknowledged by Public Health officials. The political reluctance to do so stems in part from the clearly implied origins of some stealth adapted viruses from the use of kidney cells from cytomegalovirus contaminated monkeys to produce live polio virus vaccines. It is imperative that the culturing and genetic analyses of stealth adapted viruses be pursued.
Keywords: Stealth adapted viruses, chronic fatigue syndrome, CFS, myalgic encephalomyelitis, polio vaccine, rhesus monkey, African green monkey simian cytomegalovirus, SCMV, renegade genetic sequence, transduction, homologous recombination, long non-coding rna, polymerase chain reaction, PCR, BLAST, introgression, viroids, Covid-19
CFS, chronic fatigue syndrome; CPE, cytopathic effect; CSF, cerebrospinal fluid; HHV-6, human herpes virus-6; kb, kilobases; NCBI, National Center for Biotechnology Information; PCR, polymerase chain reaction; Sbjct, Subject nucleotide sequence identified as matching to a Query sequence on NCBI’s BLAST program; SCMV, African green monkey simian cytomegalovirus
It is still being debated whether in antiquity, viruses preceded or arose from more complex genomes.1 Either possibility can explain the overall similarities of many of the viral genes in various types of viruses with comparable counterpart genes in the cells that are specifically susceptible to infection by the viruses. Indeed, this similarity is required for the complex interplay between transcripts of the virus and of cellular genes that allows for virus replication and the cytopathic effect (CPE). Eukaryotic cells and viruses have, however, largely independently evolved since becoming distinct biological entities, while still retaining the capacity for inter-genomic communications. DNA sequencing studies on viruses cultured from patients with the chronic fatigue syndrome (CFS) have changed this perception. The studies point to a much more dynamic potential of genetic interchanges and to the capacity of stealth adapted viruses to transmit “renegade” cellular sequences between individuals and even between species. As described in this article, the possibility exists of the infectious spread of major genetically determined illnesses by stealth adapted viruses.
Chronic fatigue syndrome (CFS) was initially suggested in 1988 as an appropriate diagnostic term for a prevalent illness comprising the onset of persisting or recurrent episodes of debilitating physical and/or mental fatigue, which could not be otherwise explained by a known medical illness.2 The CFS diagnosis is further supported by a combination of subjective symptoms, including cognitive and autonomic nervous system impairments; migratory localized pain in the muscles, lymph nodes, and/or joints; painful throat, and recurring headaches. The name was chosen to help counter the growing assumption that the illness was caused by chronic infection with Epstein Barr Virus (EBV).3,4 The CFS term also had the effect of deemphasizing a possible infectious origin of the illness as had been raised by the occurrence of a community outbreak occurring in the Lake Tahoe region in Nevada during 1985 and 1986.5 There had been several prior outbreaks in the 20th century of similar illnesses among hospitalized patients in England, Iceland, and the United States.6 Without an objective laboratory or clinical diagnostic marker, however, it was reasoned that these outbreaks were probably being induced through psychosocial mechanisms.7
Various earlier names had and are continuing to be applied to the CFS type of illness. The names largely depend on whether the illness is considered primarily to be psychological or organic and whether it involves only the brain, both the muscles and brain, or is a disorder of the immune system. Prominent descriptive terms are systemic exertion intolerance disease8, myalgic encephalomyelitis, post-viral syndrome, chronic fatigue immune dysfunction syndrome (CFIDS), neuromyasthenia, yuppie flu, etc.6 Unexplained fatigue is a common presenting symptom in primary care medicine, often occurring in association with stressful events.9 It was, therefore, of some concern, especially to the disability insurance industry, that the subjectively defined illness would become ever more widely reported and exacerbated by delayed recovery if CFS were attributed to a transmissible infectious agent.
Two other developments coincided with the Lake Tahoe outbreak of CFS. One was the development in 1986 of a highly sensitive molecular diagnostic assay procedure.10 It is based on the ability to selectively amplify small segments of known DNA sequences. The assay soon became known as the polymerase chain reaction (PCR). It required knowledge of the DNA sequences that flanked the segment of DNA that was intended to be amplified. PCR oligonucleotide primers that will selectively bind (hybridize) to these flanking regions are then used to exponentially amplify the intervening segment of DNA. The second development was the identification of human herpes virus-6 (HHV-6) that was distinct from EBV and other known human herpes viruses.11 I chose to undertake the logical inquiry of using the PCR assay to test CFS patients for active infection with HHV-6. With very few exceptions, specific testing for HHV-6 infection in CFS patients under the medical care of Dr. Jay Goldstein,12 a prominent clinician who was treating CFS patients, did not indicate an active ongoing HHV-6 infection. There were some weak positive results when I performed the PCR assays under less stringent conditions. This approach to performing the PCR assay can enable initial gene amplification even with only partial binding of the PCR primers to regions of the virus DNA. Additional primers were tried including a set that would readily amplify segments of all known human herpesviruses. These primers yielded discernable positive PCR results in many tested CFS patients.13-15 Strikingly positive results were seen in tested blood and cerebrospinal fluid (CSF) samples from patients with more severe neurological illnesses. A positive result was also obtained on tissue from a stereotactic brain biopsy. The biopsy showed no inflammation despite the positive PCR. It did, however, show numerous foamy vacuolated cells suggestive of a possible spumavirus infection.16 Spumaviruses differ from simple retroviruses in having sets of genes between the envelop coding gene and the right sided long terminal repeat (LTR) sequences (bel genes).17 Human T lymphotropic viruses (HTLV) also have accessory genes in this region, including the tax gene.18 Primers for the tax genes of HTLV-1 and HTLV-2 were, therefore, also used in continuing PCR studies on blood samples from CFS patients. Variable but clearly positive PCR were obtained in several of the tested CFS patients.
Buoyed by the positive PCR results, a determined and successful effort was made to culture a cytopathic (cell damaging) virus from a 43-year-old-woman who had previously been hospitalized for 7 days to rule out encephalitis or meningitis. A CSF sample obtained during her admission was without cells (acellular). Yet her blood clearly tested positive in various low stringency PCR assays. A positive virus culture was obtained from this patient in late 1990 and on many repeat occasions over the ensuring four years.19 The second positive culture was obtained from a 22-year-old woman with a 4-year history of a bipolar psychosis that was initially diagnosed as schizophrenia. She had required varying levels of institutional care for her illness. She collapsed into a coma while hallucinating in late January 1991. She experienced multiple seizures while being transported by ambulance to Los Angeles County Hospital. She also had a brief cardiac arrest while in the Emergency Room from which she was resuscitated. A CSF sample was obtained by lumbar puncture shortly after her admission. It was acellular, yet yielded a positive virus culture.20 A sample of the cultured virus was sent to the Los Angeles County Public Health Laboratory but dismissed as probably being a cell line virus contaminant. A leading virologist at the Centers for Complex Infectious Diseases and Prevention (CDC) also expressed his skepticism about any virus that was also being suggested as infecting a CFS patient. The patient attending physicians attributed her remaining vegetative state to anoxic brain damage. She died several years later.
The positive viral cultures from the blood of the CFS patient and from the CSF of the comatose patient were microscopically similar yet still somewhat distinguishable in their developments of foamy vacuolated cells with prominent syncytia (cell fusion). There was also an overall similarity in the patterns of PCR products seen in agarose gel electrophoresis when using the same set of HTLV-related PCR primers.19 Both cultures showed a prominent amplified DNA band measuring approximately 1.5 kilobases (kb). There were also several similar smaller DNA bands generated from both cultures. No apparent DNA products were identifiable when the same PCR assay was used on uninfected cultured cells.
The reproducible DNA bands seen in agarose gel electrophoresis of the DNA generated in PCR assays performed on the culture from the CFS patient are shown in Figure 1. The larger band was excised from the agarose gel, cloned into pBluescript plasmids and subsequently sequenced. This showed that the banded DNA comprised two distinct products. The larger identified as clone 15-5-4 was 1,539 nucleotides in length versus a slightly smaller product that was 1,484 nucleotides long.19 Although not initially identifiable as SCMV-derived sequences, this became apparent as corresponding SCMV genetic sequences were deposited into GenBank by other investigators.21-23 Specifically, the BLASTN program24 of the National Center of Biotechnology Information (NCBI) showed that after excluding the primer sequences, there were 95% and 93% nucleotide identity respectively of the two PCR products with regions of the SCMV genome. These values far exceeded the matchings to other known primate, non-primate, or human cytomegaloviruses.
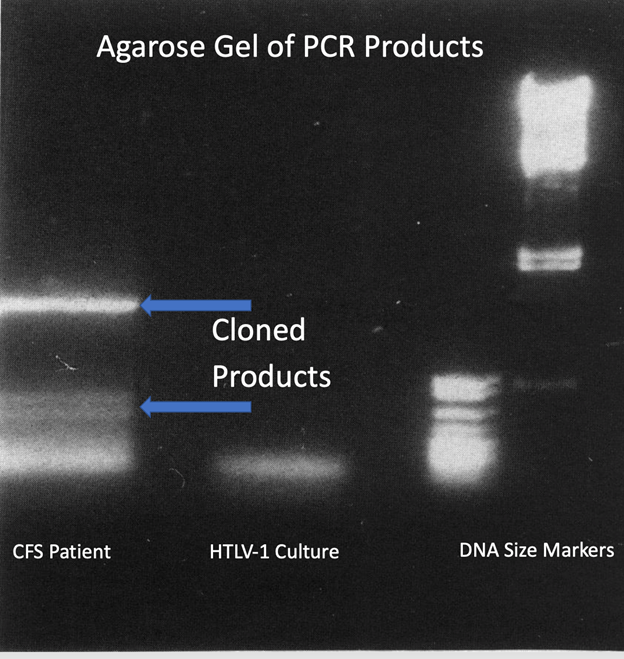
Figure 1 Ethidium bromide dye-stained DNA in an agarose gel following electrophoresis. The bands of DNA products reproducibly generated by the PCR assay performed on cells and on the filtered and pelleted culture supernatant from the stealth adapted virus culture from the CFS patient are shown in the left lane. The arrows point to the sources of DNA for cloning. The next lane indicate the PCR product obtained using the same PCR conditions on HTLV-1 infected cells. The right two lanes are DNA size markers.
The sequence on a cloned smaller band (666 nucleotide) PCR generated product did not, however, match to the SCMV genome. Yet to be fully explained, this product migrated slightly further into the agarose gel than anticipated by its size. With the completed sequencing of the human genome, GenBank assisted genetic analysis indicated that this sequence differed only slightly from a non-coding intergenomic region in the human X chromosome.25 There were 609 sequences between the two PCR primer binding sites. Of these there were 606 identical nucleotides in comparison to an intergenomic region on the human X chromosome. As shown in Figure 2, there were two single nucleotide deletions and one nucleotide substitution. The next closest matching was to a related area in the X-chromosome of chimpanzees (598 of 609 identical nucleotides). It was more distantly related to the corresponding X-chromosome of African green monkeys (553 of 609 identical nucleotides).

Figure 2 PCR Generated product matches to a region of the human X chromosome.
BLASTN matching of the sequence of one of the PCR products generated from the stealth adapted virus culture (represented as Query) with the assembled sequence of the human X chromosome (represented as Sbjct). There are three differences between the two sequences. The X chromosome sequence has an adenosine (A) nucleotide at both sequence number 30177542 and 30177538. Neither adenosine is present in the PCR product The third change is that the quinine (G) in the PCR product at nucleotide position 631 is different from the adenosine in the corresponding X chromosome sequence number 30177220.
DNA present in the larger PCR generated band from positive cultures from the CFS patient was radiolabeled and used as a hybridization probe. It hybridized with the DNA extracted from infected cells. It also hybridized with DNA extracted from the pelleted materials obtained by ultrafiltration of 0.45 m membrane filtered cultured supernatants.19 This DNA migrated in agarose gels with an apparent size of approximately 20 kb, far smaller than would be expected from an intact herpes virus. DNA extracted from the pelleted filtered supernatant and in a separate experiment, from the DNA extracted from the approximately 20 kb band were cloned into pBluescript plasmids. The T3 and T7 polymerase sites on the plasmid allowed for the insert to be sequenced from both of its ends. Of 248 either completely or partially sequenced clones, 18 have genetic sequences that closely match to cellular rather than to SCMV-derived genetic sequences.26 There are 30 clones with bacterial-derived genetic sequences.27 These bacterial sequences are not the subject of this article. The remaining clones had T3 and T7 generated sequences that could be matched to regions of the SCMV genome. It might have been expected that at least some of the inserts with cellular sequences would have most closely match to the cellular genome of African green monkey. Upon genetic analysis, however, the cellular sequences all matched more closely to sequences within the human genome. The individual matching cellular sequences are located on human different chromosomes and usually within the intron of diverse protein coding gene. These sequence data are available on GenBank and again show minor genetic differences from the originating sequences of the human genome.
PCR products were generated in several cultures of CFS patients when using the same set of PCR primers as was used in testing of the cultures from the first tested CFS patient and from the comatose patient. The PCR products generated from cultures of some of these patients included the approximately 1.5 kb DNA band along with smaller bands as were seen in the earlier PCR assays. Yet PCR products of different sizes were not uncommonly seen when using the same set of PCR primers on the positive cultures from other patients. Informative data were obtained in the sequencing of the PCR products from three such cultures.25,28-30 Only one PCR product was sequenced from the first of these three cultures (Referred to as patient LB). It showed an approximate match to a human cellular gene. Yet far closer matching, as noted by the number of matching identical nucleotides, was seen when the DNA sequence comparison was made to the assembled cellular genome of rhesus monkeys (Table 1). The GenBank Accession number of the rhesus cellular sequence, which includes the sequence matching to that of the PCR product is included in the Table.
Partial T3 and T7 generated DNA sequence data were obtained on the T3 and T7 generated end sequences of six PCR products from the virus culture of a different CFS patient (referred to in Table 1 as Patient B and C11-- clones) and on the end regions of seven PCR products from the virus culture of another CFS patient (referred to in Table 1 as patient C and as C13-- clones). The patients’ cultures were obtained at different times and the cloning studies independently performed. Yet, two out of six sequenced PCR products from the culture of Patient B (clones C1113 and C1132) and one of seven of the PCR products from the culture of Patient C (clone C13220) had all originated from the same rhesus cellular sequence that had been earlier identified in the virus culture of patient LB (GenBank Accession NC_041761.1). The matching sequences had partially diverged from each other, including slight differences between the two related sequences from the culture of the patient B. There was also genetic similarities indicative of common rhesus cellular origins of the sequences in two other clones of patient B (clones C1123 and C1163) and a clone C1313 from patient C. There was an additional shared sequence between the clones of Patients B and C (clones C1142 and C1335). Finally, while all six of the sequences in the PCR products from the second culture were of rhesus cellular origin, three of the seven sequences from the third culture matched more closely to corresponding non-coding genes in the human genome. The nucleotide identity data and the matching GenBank accession numbers are provided in Table 1.
Patient |
Clone |
Rhesus |
Human |
Species &Accession No. |
|
LB |
LB |
484/490* |
435/490 |
Rhesus |
NC_041761.1 |
B |
C1113 |
420/434 |
371/414 |
Rhesus |
NC_041761.1 |
“ |
C1123 |
358/381 |
330/369 |
Rhesus |
NC_041754.1 |
“ |
C1132 |
408/437 |
364/411 |
Rhesus |
NC_041761.1 |
“ |
C1142 |
396/411 |
114/140 |
Rhesus |
NC_041764.1 |
“ |
C1151 |
392/404 |
364/401 |
Rhesus |
NC_041760.1 |
“ |
C1163 |
191/194 |
161/180 |
Rhesus |
NC_041754.1 |
C |
C1311 |
372/425 |
412/428 |
Human |
NC_000017.11 |
“ |
C1313 |
317/329 |
295/326 |
Rhesus |
NC_041754.1 |
“ |
C1322 |
162/173 |
150/173 |
Rhesus |
NC_041761.1 |
“ |
C1325 |
266/280 |
166/186 |
Rhesus |
NC_041758.1 |
“ |
C1333 |
177/225 |
265/289 |
Human |
NC_000008.11 |
“ |
C1334 |
272/289 |
281/289 |
Human |
NC_000005.10 |
“ |
C1335 |
305/315 |
272/315 |
Rhesus |
NC_041764.1 |
Table 1 Levels of Nucleotide Identity of the Combined T3 and T7 Sequences of Cloned PCR Products Matched by BLASTN Against the Rhesus and Human Assembled Cellular Genomes
Footnote to Table: The data are from the sequencing of clones of PCR generated DNA obtained from the stealth adapted virus cultures of three patients. The patients are designated LB (Clone LB); Patient B (Clones C11 series); and Patient C (Clones C13 series). The sequences of the clone were matched against the assembled rhesus monkey and human genomes. *The highest of the two matching ratios for each clone is shown in bold typeface. Also printed is the NCBI Accession Number of the rhesus or human GenBank submission that contained the sequence, which best matched to the sequence of each clone.
Stealth adaptation was proposed as the loss of effective immune recognition of a virus due to the deletion or mutation of the genes coding for the relatively few virus components that are normally targeted by the cellular immune system.15,19-23 This adaptation can explain the lack of inflammation seen in virus infected patients31 and in virus inoculated animals.32 The incorporation of additional genetic elements may be required by certain stealth adapted viruses to regain their infectivity. Clearly, some of the incorporated additional sequences in the viruses under study are from cellular genomes. A possible mechanism for this occurring is for various stretches of transcribed cellular RNA to have crosslinked different fragments of the remaining originating virus genome. This would then lead to the incorporation of the crosslinking cellular sequences into the reformed viral genome by reverse transcription. This process can be perceived as either the hijacking of the cellular sequence by the virus or that the cellular sequences have become renegade in achieving an existence beyond their normally restricted cellular location.
Unlike most of the incorporated bacterial sequences, the cellular derived human and primate sequences so far identified in cultures of stealth adapted viruses are non-coding. The amounts of non-coding RNA in a cell exceeds that of protein coding messenger RNA.33 This may explain why only non-coding sequences have so far been identified. Based on their lengths (> 200 nucleotides), if they are incorporated as RNA, they would comprise portions of long non-coding RNA molecules. Whether the incorporated sequences contribute specific metabolic functions to the infected cells needs to be determined. There is at least some degree of genetic variability between similar originating cellular sequences either in different viruses or even within the same virus. This would argue against a tightly regulated sequence-specific function of the incorporated sequences. To this date, there are no ascribed functions of the originating long non-coding RNA cellular sequences, which have been identified as being apparently involved in the formation of stealth adapted viruses. Aberrant biological functions are, however, being increasingly ascribed to the altered expression of specific long non-coding RNA molecules. These include malignancy, degenerative illnesses, psychiatric disorders, diabetes, and various immune disorders.33
The best studied stealth adapted virus originated from SCMV. It might have been expected, therefore, to contain African green monkey cellular sequences. Instead, its cellular sequences are of human origin. Based upon the presence of both rhesus and human cellular sequences in the PCR amplified products generated from one of the other cultures, it is likely that genetic exchange, probably through homologous recombination,34 explains the replacement of original primate sequences with matching human sequences. The SCMV-derived stealth adapted viruses can be cultivated in cells of multiple species.19 By doing so, the cultivated SCMV-derived virus can be examined for the potential replacement of the human-derived cellular genes with those of the species of the infected cells. Additionally, clinical experience is consistent with stealth adapted virus spread from infected cats to humans.35 This could be a source of feline cellular sequences introgressed into infected humans.
The prototype SCMV-derived stealth adapted virus comprises multiple segments that are approximately 20 kb long. This is about the size limit for viral RNA replication by either RNA polymerases or RNA dependent DNA polymerases. Direct evidence for the involvement of RNA in the replication mechanism of at least certain stealth adapted viruses was provided by the requirement of an initial reverse transcriptase in the PCR detection of an SCMV-related stealth adapted virus in the CSF of a symptomatic healthcare provider.36 RNA based replication by either RNA polymerase or RNA dependent DNA polymerase is consistent with the apparent genetic instability of the originating viral and the incorporated cellular sequences in the stealth adapted viruses for which there are sequence data.
The very similar PCR reactivity patterns indicates that the stealth adapted virus isolated from the original CFS patient is related to the virus isolated from the patient with the severe psychotic illness. The former virus is available from the American Type Culture Collection (ATCC). Positive virus cultures were regularly obtained from inpatients at both the Los Angeles County Psychiatric Hospital and at a Community Psychiatric Hospital in Rosemead, California. There is a need to extend the sequencing on the archived virus and to culture and to sequence stealth adapted viruses from additional psychiatric patients. Possible sequence differences in the incorporated cellular genes may relate to the type and severity of the resulting illness. Yet there have been examples of CFS and a psychotic illness occurring among different members of the same family. The younger brother of the woman described in this article with the severe psychotic illness was described by his mother as being extremely lazy. The mother of a stealth adapted virus culture positive young women with schizophrenia also reported her own diagnosis as CFS. A similar circumstance occurred in another family. CFS is an imprecisely defined illness37 and is likely to reflect only a small spectrum of illnesses caused by stealth adapted viruses.
The woman with CFS from which the first stealth adapted virus isolate was obtained had rented an apartment to someone who had become her non-sexually intimate friend. This gentleman was HIV positive yet had many of the same cognitive and fatigue inducing disorders as did the woman. She, therefore, attributed her acquiring the illness from the gentleman. On several occasions, stealth adapted viruses have been cultured from HIV positive individuals and these viruses may contribute to the severity of the illness.
The sharing of related rhesus monkey cellular sequences with the other three CFS patients provides support for the possible presence of only a limited number of widely disseminated originating stealth adapted virus. The opposing viewpoint is that stealth adaptation is a generic process that can occur with all known human and animal viruses. Again, this issue can be resolved with further sequencing of stealth adapted viruses from patients with multiple illnesses and to further examine for possible correlations between the manifested disease and cell-derived genetic sequences. There is clearly the potential through the incorporation of pathogenic cellular sequences for various genetically based disorders to become infectious.
The extent to which the originating virus sequences can be deleted from stealth adapted viruses has yet to be studied. Conceivably, the transmitted cellular sequences, possibly along with transmitted bacteria-derived sequences, could become autonomous replicating elements. Indeed, the isolated non-coding cellular sequences could take the form of viroids as occur in plants.38-40 This does not apply to the prototype SCMV-derived stealth adapted virus since there are still genetic sequences corresponding to approximately half of originating SCMV genome in the more than 200 clones on which sequence data have been obtained. Interestingly, there is a very uneven distribution of the matching clones with some regions predominantly represented. Yet the original virus and cellular-derived sequences were all detected within the isolated 20 kb band of DNA.
Another concerning issue is the potential interaction between stealth adapted viruses and other viruses. The existence of stealth adapted viruses has yet to be acknowledged by Public Health officials. The reluctance to do so stems largely from the clearly implied origins of some of these viruses from the use of kidney cells from cytomegalovirus contaminated monkeys to produce live polio virus vaccines. The long covid syndrome has features of CFS and it would be worthwhile to specifically test these patients for stealth adapted viruses.41,42 Similarly, vaccine enhancement of cellular immunity against some of the remaining, yet normally not immunologically recognized components of a preexisting stealth adapted virus, may account for some of the vaccine side effects experienced by certain patients.
Genetically unstable cellular sequences are components of the stealth adapted virus that was repeatedly cultured from a patient with the chronic fatigue syndrome (CFS). Although the virus was derived from an African green monkey simian cytomegalovirus (SCMV), the cellular sequences were of human origin. One of these sequences that matched closely to an intergenomic region of the human X chromosome could be amplified by the PCR assay performed on infected but not on uninfected cells. This sequence was also likely to have been amplified in the PCR assay performed on other stealth adapted virus cultures, including the culture from a comatose woman with a four-year history of a severe psychotic illness. Rhesus monkey-derived cellular sequences were detectable in the cultures that were independently obtained from three other CFS patients. There was sharing, although not exact identity, between certain of the amplified sequences in these three cultures. In one of the cultures, while four of the amplified cellular sequences were of rhesus origin, three of the sequences were derived from the human genome. This finding is consistent with the progressive homologous recombination or exchange of primate sequences with those of the species being infected. Stealth adapted viruses can potentially transmit pathogenic cellular genetic sequences between individuals and convert certain genetic based diseases into infectious illnesses. It is important to extend the culturing and genetic sequencing of stealth adapted viruses in patients with a wide range of illnesses, including psychiatric, degenerative, and malignant diseases. The potential interaction of stealth adapted viruses with the Covid-19 virus should also be explored.
The work was supported by MI Hope Inc., a non-profit public charity.
None.

©2022 Martin. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.