Journal of
eISSN: 2373-6453


Review Article Volume 3 Issue 1
Executive Scientific Advisor at hVIVO Group plc, UK
Correspondence: Rob Lambkin-Williams, Executive Scientific Advisor at hVIVO Group plc, 42 New Road, London, E1 2AX, UK, Tel 1273915039, 07710577019
Received: December 17, 2015 | Published: January 4, 2016
Citation: Lambkin-Williams R, Mann AJ, Gilbert AS (2016) Re-Inventing the Common Cold Institute for the 21st Century. J Hum Virol Retrovirol 3(1): 00075. DOI: 10.15406/jhvrv.2016.03.00075
In this article we describe the history of Human Viral Challenge Studies, their utility and their major contribution to the development of treatments to major unmet medical needs. In the early part of the Second World War the American Red Cross-Harvard Hospital had been brought across the Atlantic as prefabricated wooden units. After assembly, the hospital opened in 1941 to deal with epidemics of infectious disease that might occur, but thereafter it had been in disuse. In 1946 it was repurposed as the Common Cold Unit also known as the Common Cold Institute; Scientists and doctors examined volunteers in quarantine, closely observing and monitoring the effect of common cold viruses that they deliberately infected them with. They were able to isolate and grow viruses in the onsite laboratories and then administer the viruses to volunteers through nasal drops.
The Unit shut in the 90’s, but experimental infection studies continued, with small motels and hotels in the USA substituting for the wooden huts on Salisbury Plain. Such studies contributed to the significant development of the new neuraminidase inhibitors during the 1990s. In 2001 we began to develop a series of well-characterised virus stocks. Here we describe the details of the model, our new purpose built unit, and its use for identifying novel antiviral compounds, novel vaccines and immunomodulators.
Keywords:HVC, Influenza virus, Common cold virus, RSV, Symptoms, H3N2, CD4+, T cells
HVC, Human Viral Challenge; MRC, Medical Research Council; CCU, Common Cold Unit; CRC, Clinical Research Centre; RSV, Respiratory Syncytial Virus; HAI, Hemagglutination Inhibition Assay
On the 21st of June 2001 we inoculated our first volunteer with influenza virus, since then we have inoculated over 2000 volunteers with three different respiratory viruses.
Over two centuries ago, Edward Jenner performed the first documented Human Viral Challenge (HVC) study with smallpox on the 14th of May 1796,1 since then the utility of such studies has been apparent. In 1931 Sir Christopher Andrews returned from the US where he had observed the use of chimpanzees in the study of influenza. However, as his return coincided with the great depression, funding for similar work in the UK was extremely limited. Sir Christopher therefore decided to enroll students from St Bartholomew’s Hospital. He explained to them that as he could not get chimpanzees, he considered the next best thing would be a “Bart’s” student. Despite the comment that “they were cheaper than chimpanzees”, over 100 students immediately enrolled, however the students had to continue their studies and were not isolated in the same way the chimpanzees had been in the USA.2 This confounded any analysis of the data as the investigators could not be certain that the symptoms were not due to any other respiratory viruses acquired in the community. The UK’s Medical Research Council (MRC) terminated the work just a year later. It was clear that the volunteers needed to be quarantined from the general population to both protect members of the public, but also to prevent volunteers acquiring a community acquired infection which would confound the study data.
In the early part of the Second World War the American Red Cross-Harvard Hospital had been brought across the Atlantic as prefabricated wooden units. After assembly, the hospital had opened in 1941 to deal with epidemics of infectious disease that might occur, but thereafter it had been in disuse. After the conclusion of the war, the virologist Sir Christopher Andrews, who first isolated the influenza virus with Patrick Laidlaw and Wilson Smith, saw it as being ideal for housing inoculated volunteers who could be isolated from each other and the community. The site was considered the ideal location to carry out such research in the isolation of the Salisbury countryside, near historic Stonehenge. Scientists and doctors could examine volunteers in quarantine, closely observing and monitoring the effect of colds. They were able to isolate and grow viruses in the onsite laboratories and then administer the viruses to volunteers through nasal drops. The first volunteers to be inoculated arrived in July 1946.
David Tyrell,2 one of the External Scientific Staff of the Medical Research Council (MRC), was asked in 1954 by the Secretary of the MRC, Sir Harold Himsworth, whether he would be willing to go to the Common Cold Unit (CCU), in Salisbury, Wiltshire, and attempt to grow the common cold virus.
David Tyrell2 took over the CCU in 1957. The unit housed healthy volunteers in relative isolation from other people, thereby reducing the risk of contact with natural sources of infection or of passing on the virus to members of the public. During its time the unit attracted 20,000 volunteers until its closure in 1989.
Tyrrell significantly advanced public understanding about the common cold. He first discovered that the common cold was caused by viruses, and that there was not a single common cold virus, but that instead, colds were caused by many different viruses.
The unit provided the breakthrough, and in January 1960 exciting papers describing the success of isolating and growing rhinoviruses, as they became known, were published in The Lancet. The notion of "the common cold virus" was dispelled as, subsequently, more than 100 different rhinovirus types were discovered, they were far from common. Other completely different cold-causing viruses called coronaviruses were also discovered. Not surprisingly, Dr Tyrrell2 gained a worldwide reputation, as did the unusual volunteer set-up.
In 1967, Tyrrell moved to head the Division of Communicable Diseases at the MRC's Clinical Research Centre (CRC), built in association with Northwick Park Hospital in Harrow, Middlesex. He was appointed Deputy Director of the CRC in 1970 but visited the CCU regularly, maintaining control of the work there.
He once said, "People say that place never found the cold cure, did it?" Of course, the enlightened realized that the finding of so many different viruses made the possibility of an effective vaccine hopeless. So far as David Tyrrell was concerned, there could be no self-recrimination. His was a star-studded career, about which the majority of research investigators can only dream. At the CCU, Tyrrell ran an on-going program of volunteer trials alongside laboratory work, in order to examine the clinical characteristics of the common cold and gain an understanding of the mechanics of how colds are transmitted.
The unusual set up of the trial received substantial media attention, and the CCU was often discussed on radio and TV. Through press releases and interviews, the staff of the CCU pitched the trial as an ideal budget holiday, claiming it was ‘the best package holiday anywhere. News outlets described the trials as a ‘holiday not to be sneezed at’, with ‘cold comfort’ provided. As a result of this enthusiasm, the program was often over-subscribed, and volunteers were queuing up to take part in the unique trials. Many of the CCU volunteers took part more than once, and some even went on honeymoons to the site, or used it as an opportunity for quiet study in the CCU library.
Post 1989 experimental infection studies continued, with small motels and hotels in the USA substituting for the wooden huts on Salisbury Plain. Such studies contributed to the significant development of the new neuraminidase inhibitors during the 1990s.3-13 In 2001 we began to develop a series of well-characterized virus stocks. With the Human Viral Challenge Model gaining acceptance in an increasing number of peer-reviewed publications, we designed and built our own dedicated, bespoke quarantine unit in London, which opened in early 2011. Before this we used university halls of residence and hotels.
We have, as others have,3,7,14,15 used the model to;
The FDA provided guidance in 2011 on the use of Human Viral Challenge Studies; “Phase 2A: Challenge trials after initial antiviral activity assessments and phase 1 human pharmacokinetic (PK) and tolerability trials, several sponsors have conducted challenge trials. In challenge trials, healthy volunteers are administered an investigational antiviral drug either before (prophylaxis trials) or after (treatment trials) inoculation with the established challenge strain of influenza virus. Challenge strains are influenza viruses that produce a milder set of symptoms compared to naturally occurring influenza. Pharmacodynamic (PD) endpoints in challenge trials include clinical respiratory symptoms, nasal discharge weight, and quantitative measurements of viral shedding in nasal washes. Sponsors are encouraged to include assessments of resistance in their challenge trials.
Challenge trials can provide useful exposure-response and safety information, as well as an opportunity to demonstrate pharmacological antiviral activity in humans under controlled conditions outside the influenza season. Data from challenge trials can contribute to dose selection for phase 2B and phase 3 trials, and provide the opportunity to explore the effects of different times of drug initiation relative to virus exposure.….”
The HVC model using healthy volunteers provides a unique opportunity to describe the viral lifecycle as: the time point of infection is known with certainty, nasal virus shedding can be measured, symptoms are recorded prospectively and participants are selected with low antibody titres to ensure a statistically significant infection rate with a relatively small number of volunteers.16,17
The outcome of any study will depend on three factors: the pathogenicity of the virus used to challenge volunteers, the host status as regards pre-existing homo- and hetero-subtypic immunity, and how the experimental challenge model accurately reflects transmission of influenza in the real world. It has been suggested, as per the FDA statement above, that the viruses used in experimental studies were of moderate pathogenicity by comparison with wild-type seasonal influenza viruses (15, 24, 47, 48).
It is noteworthy that two (8 percent) of 24 studies using A/H3N2, versus 19 (76 percent) of 25 studies using A/H1N1, were published after 1990 (p < 0.001), indicating a trend in the use of these respective influenza virus subtypes. This trend was likely due to the opinion that A/H3N2 infections gave more severe illnesses than did A/H1N1 infections, as higher rates of mortality or hospitalization have been reported with the A/H3N2 subtypes (49-51). However, no arguments supporting the idea that the challenge viruses were only moderately pathogenic could be supported. In particular, viral shedding and illness proportions did not differ according to the virus subtype. Two recent studies used the pandemic H1N1 strain to challenge seronegative volunteers (46, 52). They reported a proportion of ‘‘any illness’’ of 60 percent (15 of 25), consistent with the summary measure. Our first study in influenza was conducted in 2001 and involved eight volunteers who were infected with a B virus in a hall of residence at the University of London (Figure 1).
We chose a B virus, which had been manufactured under GMP conditions with limited cell passage, as we felt this was the safest first step in the re-invention of the Human Viral Challenge Model. In groups of two volunteers, we inoculated four different virus titres to determine the optimal titre to use in a subsequent study. Although the groups were small, the data was good and a “dose” response observed. We chose a titre of 105 TCID50 and then conducted a study on a vaccine; although the vaccine was not effective, we did observe a good infection rate and viral pathogenicity. Since then we have conducted multiple studies and over 2000 volunteers have been inoculated with Influenza A and B, Respiratory Syncytial Virus (RSV) or Human Rhinovirus (HRV), and have shown multiple proof of concept.18,19
A complete, GMP, manufactured inoculum virus panel has been built, including three influenza H3 viruses, two influenza H1 viruses, two HRV virus and a single RSV virus. Importantly, we have extensive historic data on these viruses which allows thorough powering calculations and end point determination for future studies. Normally study sampling is defined based around an endpoint of infection rate, virus shedding or symptoms. In addition where needed, we have data where the sample size can be calculated based on a novel biomarker, such as IL-8, we have done this successfully previously.
A volunteer on the very first study commented recently; “Flu camp was a brilliant experience (even though we were essentially being shut off from the outside world for a week to be given, potentially, the flu!). I ended up taking part twice because I loved it so much. I was part of the very first trial and the name 'flu camp' was born because it was like being in a holiday camp with much fun which came out of bonding with fellow 'inmates'. Really well organized and we were all made to feel very valued and well looked after. Never a dull moment.“ We have used the model to investigate three key respiratory viruses; influenza, Human Rhinovirus and Respiratory Syncytial Virus.
Influenza
Influenza and its associated diseases are a major cause of morbidity and mortality.20 It is important to note that Influenza A (H3N2) causes the greatest morbidity and mortality on an annual basis21 even when compared to the recent 2009 pandemic H1N1 strain;22 hence it is an important focus for model development and antivirals, vaccines and novel diagnostics.
As a slightly flippant aside, using the data from over a decade, we recently compared the difference between the sexes, do men really get sicker than women, do they just complain more, or are they being given a rough ride by their other halves? We compared the frequency and severity of disease, how much virus the person produced and their immunity. Interestingly, there was no difference between men and women in either how sick they were or how much virus replicated. Simply, man flu is a myth!
However, what is not flippant, is how serious respiratory viral infections can be in children, the elderly and at-risk groups such as asthmatics or those with other chronic underlying conditions. How well the flu vaccine works can range from season to season and is dependent on the similarity or "match" between the circulating flu viruses and the influenza strains in the flu vaccine itself, as recommended by the World Health Organisation (WHO) each year using surveillance-based forecasts as to which viruses are most likely to cause illness in the coming season.
Studies show that current vaccination reduces flu-related hospitalisations among adults of all ages by 70% and even more in those over 50 years of age. Vaccination helps protect women during pregnancy and their babies for up to 6 months after they are born. Estimates of the annual number of deaths attributable to flu in the UK vary with an average of around 8,000 per year. Flu vaccination is strongly recommended for those in at risk groups such as asthmatics, those with chronic health conditions and those over 65. Although the current flu vaccine is not perfect, the overall evidence supports the public health benefit of vaccination.
Based on the recommendations from the WHO, last year’s flu vaccine (2014-2015) production started in the second quarter of the year ready for the vaccination campaign in the third quarter - it takes six months to manufacture the vaccine. However, the virus selected for the vaccine against the most serious strain (H3) did not match the virus circulating that winter- there was a mismatch. The vaccine was estimated to work in just 3% of cases, compared with typical past effectiveness.
According to the CDC, more than two-thirds of Influenza A (H3N2) viruses in circulation were mismatched from the vaccine. UK based studies confirmed this. Unfortunately it is believed that the mortality rate in the UK due to the mismatch of the vaccine increased by up to 50%. What this tells us is that when the flu vaccine is properly matched, it is effective in saving lives and therefore those in at risk groups and children should be encouraged to get the vaccine. However when the vaccine is mismatched more people die. The need for a better influenza vaccine is clear, in the meantime the current vaccine does save lives. The utility of the Human Viral Challenge model in evaluating these new vaccines, and how such data can influence the subsequent field based studies, is vital.
The HVC model allows us to investigate the effectiveness of new vaccines without relying on the old paradigm of anti HA antibodies as the correlate of protection.23-25 We recently chose an H3N2 influenza subtype to manufacture, rather than H1N1, given that this strain has the most substantial impact in terms of morbidity or mortality annually as described by the Centre for Disease Control.26 We first subjected the virus batch to rigorous adventitious agent testing, confirmed the virus to be wild-type by Sanger sequencing and determined the virus titres appropriate for human use via the established ferret model. We built on our previous experience with other H3N2 and H1N1 viruses to develop this unique model.
We conducted an initial safety and characterisation study in healthy adult volunteers, utilising our unique clinical quarantine facility in London, UK. In this study we demonstrated this new influenza (H3N2) challenge virus to be both safe and pathogenic with an appropriate level of disease in volunteers. Furthermore, by inoculating volunteers with a range of different inoculum titres, we established the minimum infectious titre required to achieve reproducible disease whilst ensuring a sensitive model that can be translated to design subsequent field based studies.
Human Rhinovirus
Human Rhinovirus (HRV): This “common cold virus” is generally thought to cause a mild illness. However, in asthmatics it can cause serious illness. The cell based immune response is sub divided into Th1 or Th2-based responses. Infection with respiratory viruses has been shown to be a problem in asthmatics as these two sides of the cell based immune system may be unbalanced. We have conducted studies with HRV to investigate this in depth, using the Human Viral Challenge Model.
An HVC study conducted by us demonstrated the efficacy of a small molecule and allowed for the successful design and implementation of a field study in asthmatics.
Respiratory Syncytial Virus
Respiratory syncytial virus (RSV): RSV is of serious concern in infants; almost all infants will become infected by the age of 2. The virus is a major cause of lower respiratory tract infection, which in some is so severe it requires admission to hospital and intensive care. In the worst cases the virus is fatal resulting in thousands of infants dying globally annually.
RSV is generally thought of as a risk to children, but the infection in the elderly is also serious and is generally overlooked; mortality from this virus is second only to influenza in this.27
The only approved therapy for RSV infection at present is ribavirin which is rarely used in European or US paediatric populations. Ribavirin has limited efficacy and unfavorable toxicity. The only other licensed product is the monoclonal antibody, Palvizumab, which is only partially effective, can only be used prophylactically and covers a small proportion of the at risk population.28,29
“The advantages of a safe, reproducible human model are incalculable. This model permits the relatively quick and efficient study of new therapeutics in humans and assists in making critical decisions whether to advance a product into costly human trials in populations at highest risk for disease; children, elderly or immunocompromised patients. This constitutes a major and welcome advance in the field of RSV.30
There are no appropriate antiviral therapies available for RSV and no vaccines. Indeed, an experimental vaccine tested during the sixties in children caused exacerbation of disease - some of the children in the clinical study died. A vaccine is desperately needed!
The Importance of a Standardized Diary Card to Compare the Pathogenicity of Influenza, Respiratory Syncytial Virus and Rhinovirus in the Human Viral Challenge Model
We have been inoculating volunteers intranasally with clinical strains of either Influenza A/Wisconsin/67/2005 RSV A Memphis 37(b) or HRV 16 at our high containment quarantine facility in Whitechapel, UK. In each case viral loads were determined by quantitative culture methods (either pfu/mL for RSV or TCID50/mL for flu and HRV), symptoms were measured using a standardized symptom score diary card (which has been used in multiple studies over 15 years) and mucus secretions were determined by collection of tissues once daily. From the data it is apparent that in each case the viral load is closely correlated with the level of symptoms reported, suggesting that the level of illness seen is directly linked to the amount of viral shedding that is ongoing within a subject. Mucus weights also appeared to be a highly accurate indicator of the level of illness. Furthermore, breaking down the types of reported symptoms for each virus into ‘Upper Respiratory’, ‘Lower Respiratory’ and ‘Systemic’ categories also yielded several interesting comparisons between the viruses, with level of categorical symptoms reported for RSV and HRV being directly comparable contrasting with a higher proportion of symptomology being systemic for influenza.
Taken together, these findings reveal the disease dynamics observed in these three viruses, which have been demonstrated to all have different durations and intensity of viral shedding. Ultimately this highlights the value of the HVC model in interpreting the comparative pathogenesis of the three most important seasonal respiratory viral infections. Importantly, the use of a standardized diary card uniquely developed by our group and used consistently over 15 years has allowed this comparison (Figure 2).
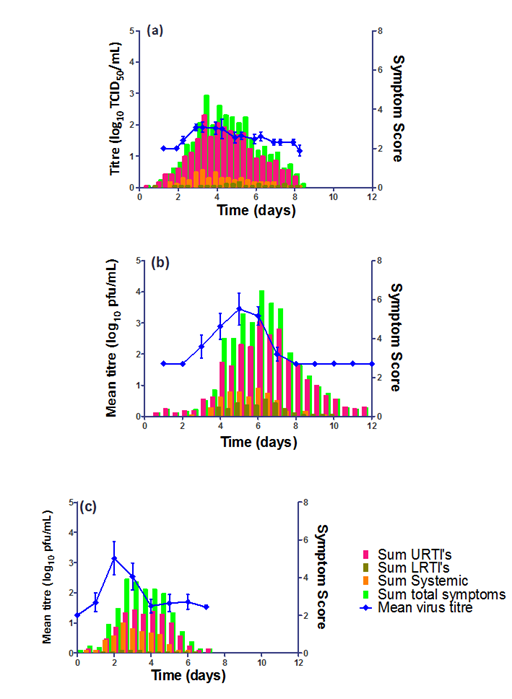
Figure 2 Temporal comparisons of the mean sum symptomology and virus titre for (a) HRV, (b) RSV and (c) Influenza H3N2 over a typical quarantine period. Note that the standard error of the mean (SEM) values for the virus tires are shown in dashed blue lines above and below the mean value.
Using the HVC Model to Evaluate Novel Vaccines
The Human Viral Challenge model is also important in analyzing novel vaccines. For example, we used the model to determine the efficacy of a novel Proteosome-adjuvanted trivalent inactivated influenza vaccine (P-TIV) administered intra-nasally. The vaccine was shown to be effective, safe, well tolerated, immunogenic - in both systemic and mucosal compartments - and effective at preventing influenza illness.
In two separate studies using the HVC model, subjects were selected for susceptibility to A/Panama/2007/1999 (H3N2) virus and then dosed with one of three regimens: inactivated vaccine (A/New Caledonia/20/1999 (H1N1), A/Panama/2007/1999 (H3N2), B/Victoria/504/2000 or B/Shangdong/7/1997) or placebo via a nasal spray. One or two doses were given, 14 days apart, before subjects were challenged with ~8.5 x 105 EID50 of A/Panama/2007/1999 (H3N2) virus. Immune responses to the vaccine antigens were measured, namely serum IgG (via the haemagglutination inhibition assay (HAI)) and nasal wash secretory IgA (sIgA) antibodies (via ELISA). Viral titres in nasal washes and symptoms of influenza illness were assessed after viral challenge and compared.
Vaccine reactogenicity was mild, predictable and generally consistent with earlier Phase 1 safety studies with this vaccine. Seroconversion to A/Panama/2007/1999 (H3N2) occurred in 57% to 77% of subjects in active dosing groups and 2% of placebo subjects. The greatest geometric mean fold rise in sIgA (post-vaccination (but prior to challenge) sIgA over pre-vaccination sIgA) to A/Panama/2007/1999 (H3N2) was observed in the groups that had been vaccinated twice. Immune responses to H1N1 were comparable to those for H3N2 and lower for B strains. Mean fold rises in sIgA were significant for all strains.
Intranasal vaccination significantly protected against laboratory confirmed illness following challenge with A/Panama/2007/1999 (H3N2). Data from both studies were pooled and vaccine efficacy calculated against rates observed in placebo recipients. Efficacy between active dosing groups ranged from 58% to 82% for any influenza illness with laboratory confirmed infection (as defined by seroconversion following inoculation), 67% to 85% for systemic or lower respiratory illness and laboratory confirmed influenza, and 65% to 100% for febrile illness and laboratory confirmed influenza. The two dose regimen was superior to the one dose regimen.
Using the HVC Model to Develop a Universal Influenza Vaccine
An important utility of the HVC model is its ability to dissect the correlates of immune protection against respiratory viruses. Protective immunity against influenza virus infection is mediated by neutralizing antibodies, but the precise role of T cells in human influenza immunity is uncertain. We conducted studies in healthy volunteers with no detectable antibodies to the challenge viruses H3N2 or H1N1. We mapped T cell responses to influenza before and during infection. We found a large increase in influenza-specific T cell responses by day 7, when virus was completely cleared from nasal samples and serum antibodies were still undetectable. Pre-existing CD4+, but not CD8+, T cells responding to influenza internal proteins were associated with lower virus shedding and less severe illness. These CD4+ cells also responded to pandemic H1N1 (A/CA/07/2009) peptides and showed evidence of cytotoxic activity. These cells are an important statistical correlate of homotypic and heterotypic response and may limit severity of influenza infection by new strains in the absence of specific antibody responses. Our results provide information that may aid the design of future vaccines against emerging influenza strains.31
As a further example, the importance of the Human Viral Challenge model is demonstrated by a study we recently conducted, along with colleagues at Oxford University, where we successfully tested a novel influenza vaccine designed to boost the cellular side of the immune system.32 The key thing is that other parts of the immune system may give a broader protective immune response, which could last over several seasons (intra-seasonal) and importantly may provide immunity against any new pandemic viruses, including bird flu.
Using the HVC Model to Discover New Treatments for Asthma and COPD
Human Rhinovirus infection is an important precursor to asthma and chronic obstructive pulmonary disease exacerbations and the HVC model provides a powerful tool in studying these and other chronic respiratory diseases. A HRV-16 isolate from an 18-year-old experimentally infected healthy female volunteer (University of Virginia Children’s Hospital, USA) was obtained with appropriate medical history and consent. We manufactured a new HRV-16 stock by minimal passage in a WI-38 cell line under Good Manufacturing Practice conditions. Having first subjected the stock to rigorous adventitious agent testing and determining the virus suitability for human use, we conducted an initial safety and pathogenicity clinical study in adult volunteers in our dedicated clinical quarantine facility in London.
In a recent study we have demonstrated the new Wild-Type HRV-16 Challenge Virus to be both safe and pathogenic, causing an appropriate level of disease in experimentally inoculated healthy adult volunteers. Furthermore, by inoculating volunteers with a range of different inoculum titres, we have established the minimum inoculum titre required to achieve reproducible disease. We have demonstrated that although inoculation titres as low as 1 x TCID50 can produce relatively high infection rates, the optimal titre for progression with future HRV challenge model development with this virus stock was 10 x TCID50. Studies currently underway are evaluating the use of this virus as a challenge agent in asthmatics.
Using the HVC Model to Understand Correlates of Protection
In addition, we have been able to use the HVC model to better understand the correlates of protection against influenza, which are poorly understood. However, in the one study, we confirmed that protection against influenza infection was significantly correlated with the pre-challenge titres of serum IgG (measured by the HAI assay) (p=0.0003) and mucosal sIgA (as measured by ELISA) (p=<0.0001) individually, and serum IgG (p=0.028) and sIgA (p=0.0014) together. Levels of serum IgG and sIgA were inversely related to rates of influenza infection and illness (Figure 3).
Using the HVC Model to Evaluate Novel Immunotherapies
We have used the HVC model to evaluate novel immunotherapies, for example anti-M2e a human monoclonal antibody targeting a conserved epitope on M2e, was explored.33 Healthy volunteers were inoculated with influenza A/Wisconsin/67/2005 (H3N2) and received a single dose of the the investigational medicinal product (IMP), or placebo 24 hours later. Subjects were monitored for symptoms, viral shedding, and safety, and also cytokine measurements. The IMP-treated subjects showed a 35% reduction (P = 0.047) in median total symptom area under the curve (days 1-7) and 2.2 log reduction in median viral load area under the curve (days 2-7) by quantitative polymerase chain reaction (P = 0.09) compared with placebo-treated subjects. The monoclonal antibody was safe and well tolerated with no additional safety signals after administration of oseltamivir. Serum cytokine levels (interferon γ, tumor necrosis factor α, and interleukin 8 and 10) were similar in both groups. These data indicate that the monoclonal antibody may provide immediate immunity and therapeutic benefit in influenza A infection, with no apparent emergence of resistant virus. The monoclonal antibody was safe with no evidence of immune exacerbation based on serum cytokine expression.
Using the HVC Model to Evaluate Novel Anti-Viral Agents
As an example, using the HVC model we conducted a double-blind, placebo-controlled study of an oral RSV-entry inhibitor in healthy adults who were challenge with of RSV. Participants were monitored for 12 days, in this study however we waited until we detected RSV shedding in the nose using qic PCR.17 At the time of a positive test for RSV infection or 5 days after inoculation, whichever occurred first, participants were randomly assigned to receive the IMP or placebo in one of seven sequential cohorts. Cohorts 1 to 4 received a first dose of 50 mg of IMP and then 25 mg daily for the next 4 days, cohort 5 received a first dose of 50 mg and then 25 mg daily for the next 2 days, cohort 6 received one 100-mg dose, and cohort 7 received a first dose of 10 mg and then 5 mg daily for the next 4 days. Dose selection for cohorts 5, 6, and 7 occurred after an interim analysis of data for cohorts 1 to 4. The primary end point was the area under the curve (AUC) for the viral load, which was assessed after administration of the first dose through the 12th day after inoculation. Secondary end points were mucus weight and symptom scores.
Among the 54 participants in cohorts 1 to 4 who were infected with RSV, active treatment was associated with a lower viral load (adjusted mean, 250.7 vs. 757.7 log10 plaque-forming-unit equivalents [PFUe] × hours per milliliter; P<0.001), lower total mucus weight (mean, 6.9 g vs. 15.1 g; P=0.03), and a lower AUC for the change from baseline in symptom scores (adjusted mean, -20.2 vs. 204.9 × hours; P=0.005). The results were similar in cohorts 5, 6, and 7. Treatment with the IMP reduced the viral load and the severity of clinical disease in a challenge study of healthy adults.
In another study to evaluate a small molecule fusion inhibitor of RSV which had a favorable safety and pharmacokinetic profile in Phase 1 studies. We utilized the HVC Model to determine efficacy, Healthy male and female volunteers were chosen that had a neutralizing titre in the lower 50% of those screened, and were inoculated in each nostril twice (total of 1 ml of 10*4 pfu) over 2 minutes with virus. An IMP or Placebo was subsequently administered orally using two dosing regimens at 48, 72, or 120 hours after intranasal inoculation. In the first dosing regimen the IMP was administered as a single loading dose of 450 mg at either 48 (Regime 1a) or 72 hours (Regime 1b) followed by 150 mg every 12 hours for 8 further doses. In the second dosing regimen a single 600mg loading dose was administered followed by 300 mg every 12 hours for 8 further doses. Virus titre was determined by qPCR daily nasal washes were performed starting at 48 hours post inoculation as always. Symptom scores and total nasal secretions were also collected.
The virus was detectable in the majority of placebo volunteers and rose in titre with a peak value on days 5-8, this then declined over the ensuing week. The IMP administered at 48 hours (Dosing regimen 1a, Loading dose 450mg and then 150mg every 12 hours) had significant impact on cumulative nasal viral load (p = 0.035), symptoms, and nasal secretions, but had no effect when administered at 72 (Dosing regimen 1b, 450mg and then 150mg or 120 hours post inoculation (Dosing regimen 2 600mg then 300mg every 12 hours.
Using the model we showed the IMP was an effective treatment in the RSV HVC model when administered at up to 48 hours post inoculation, but did not improve virologic or symptomatic parameters when administered at 72 or 120 hours.
Using the HVC Model to Develop Novel Early Stage Diagnostics
There is great potential for host-based gene expression analysis to impact the early diagnosis of infectious diseases. In particular, the influenza pandemic of 2009 highlighted the challenges and limitations of traditional pathogen-based testing for suspected upper respiratory viral infection. We inoculated human volunteers with either influenza A (A/Brisbane/59/2007 (H1N1) or A/Wisconsin/67/2005 (H3N2)), and assayed the peripheral blood transcriptome every 8 hours for 7 days. Of 41 inoculated volunteers, 18 (44%) developed symptomatic infection. Using unbiased sparse latent factor regression analysis, we, and our colleagues,34 generated a gene signature (or factor) for symptomatic influenza capable of detecting 94% of infected cases. This gene signature is detectable as early as 29 hours post-exposure and achieves maximal accuracy on average 43 hours (p = 0.003, H1N1) and 38 hours (p-value = 0.005, H3N2) before peak clinical symptoms. In order to test the relevance of these findings in naturally acquired disease, a composite influenza A signature built from these challenge studies was applied to Emergency Department patients where it discriminates between swine-origin influenza A/H1N1 (2009) infected and non-infected individuals with 92% accuracy. The host genomic response to Influenza infection is robust and may provide the means for detection before typical clinical symptoms are apparent.
We continue to infect volunteers with respiratory viruses such as Flu, Rhinovirus or Respiratory Syncytial Virus and quarantine them in a dedicated facility for up to two weeks. We do this to determine how the virus and the human body interact, to better understand all components of the immune system. Hopefully we can help develop the most effective vaccine for the future for all three viruses and new treatments for infection. Below is a summary of our experience across all viruses (Table 1).
Virus |
# Clinical Studies |
# Inoculated |
Influenza |
26 |
1117 |
RSV |
12 |
745 |
HRV |
4 |
181 |
Total |
42 |
2043 |
Table 1 Summary our Human Virus challenge studies
We have demonstrated the utility of the HVC model in determining the efficacy of novel antivirals, vaccines and immunotherapies, whilst better understanding the correlates of protection to infection and potentially early stage diagnostics. The question is what next can the Human Viral Challenge Model can be used for?
None.
None.

©2016 Lambkin-Williams, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.