Journal of
eISSN: 2373-6453


Research Article Volume 3 Issue 5
1Department of Medical Laboratory Sciences (DMLS), Faculty of Health and Allied Sciences, Imperial College of Business Studies, Pakistan
2Imperial Post Graduate Medical Institute (IPGMI), Faculty of Health and Allied Sciences, Imperial College of Business Studies, Pakistan
3Department of Diet and Nutritional Sciences (DDNS), Faculty of Health and Allied Sciences, Imperial College of Business Studies, Pakistan
4Department of Radiological Sciences and Medical Imaging (DRSMI), Faculty of Health and Allied Sciences, Imperial College of Business Studies, Pakistan
Correspondence: Ahmed Bilal Waqar, Dean & Director IPGMI, Imperial Post Graduate Medical Institute (IPGMI), Faculty of Health and Allied Sciences, Imperial College of Business Studies, Lahore, Tel (92) 3349686443
Received: August 26, 2016 | Published: September 7, 2016
Citation: Atif M, Imran M, Qamar Z, Javaid MU, Irfan M, et al. (2016) Phylogeny of Dengue Virus 2 based upon the NS3 Gene among USA, Thailand, Singapore, Japan and Philippine. J Hum Virol Retrovirol 3(5): 00110. DOI: 10.15406/jhvrv.2016.03.00110
Dengue virus is an arbovirus belonging to family flaviviridae causes dengue fever (DF) and dengue hemorrhagic fever (DHF). Dengue virus circulates in most tropical and subtropical regions or the world with 50–100 million human cases annually. Dengue viruses have four known serotypes designated as DENV-1, DENV-2, DENV-3, and DENV-4, with each serotype can cause full spectrum of sign and symptoms associated with DF and DHF. Despite the threat of the dengue virus studies of nucleotide divergence among the different serotypes has largely been limited to a single gene. This lack of basic knowledge of viral diversity severely limits vaccine and anti-viral therapy development efforts. Previously core and pre membrane genes of Dengue have been used for phylogenetic analysis. We used NS3 gene of dengue virus, which is a conserved region of the dengue virus 2 to study the phylogeny of dengue among Philippine, Vietnam, Thailand, Japan, China, Singapore, Sri Lanka and Taiwan. Phylogenetic Analysis revealed that serotypes of Singapore, Sri Lanka, China, USA and Taiwan are likely ancestors. Serotypes of Thailand occupy an intermediate position and serotypes of Japan, Philippine and Vietnam are descendant from serotypes of Thailand.
Keywords:Dengue virus, Dengue fever, Phylogeny, Bioinformatics
DF, Dengue Fever; DHF, Hemorrhagic Fever; DENV, Dengue Virus; NS, Non-Structural; NJ, Neighbor Joining; UPGMA, Unweighted Pair Group Method with Arithmetic Mean; ML, Maximum Likelihood
Dengue Virus (DENV) belonging to family flaviviridae is a positive sense RNA virus. Falviviridae family also includes West Nile Virus, Hepatitis C Virus and Yellow Fever Virus. Four serotypes of Dengue Virus (DEN1 to DEN-4) have been isolated till now. Sequencing of dengue viral RNA has allowed us to further identify strain variation within a serotype resulting in different genotypes of dengue virus. Dengue virus is prevalent in areas of Asia, Africa, Central and South America.1
Dengue fever is affecting every population of the world with 50-100 million human cases annually (Gubler, 2002). It is recently identified that there are more than 3.6 billion people at danger for DF; more than 2 million people are at risk of DHF and more than 21,000 deaths occurring each year because of DF and DHF.2
Dengue virus is an RNA virus with a genome size of approximately 11.0 kb.3 Genome of dengue virus consists of three structural genes: the capsid (C), the membrane (M) and the envelope (E). In addition to three structural genes, dengue virus genome also contains seven non-structural (NS) genes known by the abbreviations NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5. NS3 region of the dengue genome is considered very suitable for phylogenetic analysis.4 During the past decade, DHF epidemics have occurred in India, Pakistan, Bangladesh, China, Sri Lanka and Maldives.5
First outbreak of DHF in Pakistan was reported in 1994 in Karachi.6 After that many outbreaks of DHF have reported from Pakistan.7 Chan and colleagues reported DEN-1 and DEN-2 in three out of ten tested patients for dengue virus in the first outbreak of DHF. In the following year, DEN-2 infection was isolated from the province of Baluchistan.8 Another outbreak of DHF was observed in Karachi in 2005 and Jamil and colleagues reported DEN-3 from this outbreak.9 Later Khan and colleagues reported co-infection of DEN-2 and DEN-3 in 2006 outbreak in Karachi. From Karachi Dengue virus spread to other areas of Pakistan including Lahore. Many outbreaks of DHF have been reported in Lahore including outbreaks of 2008, 2010 and 2011.
Dengue virus infection is now common in Pakistan including Karachi, Lahore, Gujrat, Rawalpindi and Multan. Hamayoun and colleagues published data from the 2008 outbreak of dengue virus in Lahore showing that out of 17 samples processed via real-time PCR, ten of their patients had DEN-4, five had DEN-2 and two had DEN-3 infection.10
Increased population, increased tourism between dengue-endemic countries, globalized markets and unplanned urbanization particularly in tropical countries has markedly influenced the epidemiology of DF. Due to the above societal shifts, all of the endemic areas have now changed into hyper endemic areas, with multiple co circulating dengue virus serotypes.11-14 How such diversity impacts virulence and greater inflammatory responses remains unanswered.
Objective
To find out the evolutionary pattern of Dengue virus 2 based on the NS3 region of dengue genome.
Sampling
The sequences were retrieved from GenBank data base and 30 diverse sequences of NS3 gene from different geographical regions were selected for serotype 2. For serotype 3, one sequence from China was selected as an outgroup. A region corresponding to nucleotide 4,758- 6621 (1864-bp) for serotype 2 was chosen as shown in Table 1.
Serial # |
Serotype Location |
Targeted Sequence |
Gene Bank Accession # |
1 |
Thailand |
NS3 |
DQ181802 |
2 |
Thailand |
NS3 |
DQ181803 |
3 |
Thailand |
NS3 |
DQ181804 |
4 |
Thailand |
NS3 |
DQ181805 |
5 |
Thailand |
NS3 |
DQ181806 |
6 |
Thailand |
NS3 |
DQ181797 |
7 |
Thailand |
NS3 |
NC001474 |
8 |
Singapore |
NS3 |
GQ398257 |
9 |
Singapore |
NS3 |
GQ398268 |
10 |
Singapore |
NS3 |
GQ398269 |
11 |
Japan |
NS3 |
AF169681 |
12 |
Japan |
NS3 |
AF169682 |
13 |
Japan |
NS3 |
AF169683 |
14 |
Japan |
NS3 |
AF169688 |
15 |
Japan |
NS3 |
AF022435 |
16 |
Japan |
NS3 |
AF022440 |
17 |
Philippine |
NS3 |
FJ639710 |
18 |
Philippine |
NS3 |
FJ639711 |
19 |
Philippine |
NS3 |
FJ639708 |
20 |
Philippine |
NS3 |
FJ639718 |
21 |
Philippine |
NS3 |
GU131932 |
22 |
Vietnam |
NS3 |
FM210215 |
23 |
Vietnam |
NS3 |
EU482774 |
24 |
USA |
NS3 |
JF730055 |
25 |
USA |
NS3 |
HQ541798 |
26 |
USA |
NS3 |
HQ541799 |
27 |
Taiwan |
NS3 |
HQ891023 |
28 |
Taiwan |
NS3 |
HQ891024 |
29 |
Sri Lanka |
NS3 |
GQ252676 |
30 |
China |
NS3 |
FJ196851 |
Out Group |
|||
31 |
China |
NS3 of dengue 3 |
GU363549 |
Table 1 Table showing the accession number of all samples including their location
Sequence alignment strategies
Phylogenetic analysis was constructed among the sequenced isolates from different geographical distribution. Multiple sequence alignment and phylogenetic analysis of the sequences were done by using Geneious and trimming of the aligned sequence was done on Bioedit software. Phylogenetic trees were constructed using the Geneious and MEGA 4 software.
Phylogenetic analysis
Trees were made by the following methods
Neighbor Joining (NJ): Tree was made on geneious by using Geneious Tree Builder. Genetic distance model used was Tamura-Nei. Tree building method was NJ method. GU363549 was selected as an outgroup. Bootstrapping was done by selecting 1000 random seeds and 100 numbers of replicate
Unweighted Pair Group Method with Arithmetic Mean (UPGMA): Tree was made on geneious by using Geneious Tree Builder. Genetic distance model used was Tamura-Nei. Tree building method was UPGMA method. GU363549 was selected as an outgroup. Bootstrapping was done by selecting 1000 random seeds and 100 numbers of replicates.
Mr. Bayes: Mr. Bayes plugin was installed from Geneious website. Tree was constructed by using model HKY85. GU363549 was selected as an outgroup. Reminder settings were default settings.
Maximum Likelihood (ML): PHYML plugin was installed from Geneious website. Tree was constructed by using model HKY85. Bootstraping was done by using number of bootsrap dataset to 100. Other settings were default settings.
Parsimony: Alignment file was exported from geneious and saved in FASTA format. This file was open in MEGA 4 and saved in Mega format. Tree was considered by MEGA4 using the Maximum Parsimony Method.
A total of 30 DEN-2 NS3 sequences were used in this study. All the sequences were obtained from the GenBank which included DEN-2 isolate from Thailand, Singapore, Japan, Philippine, USA, Vietnam, Taiwan, Sri Lanka and China.
The results of all the methods were comparable apart from minor variations. Four clades were obtained by NJ, UPGMA and Parsimony methods (I , II, III and IV) as shown in (Figure 1,2 & 5) Three clades were obtained by Mr. Bayes method and two clades were obtained by the Maximum Likelihood method as shown in (Figure 3 & 4). Represented serotypes of all the clades were almost similar in all the five building methods.
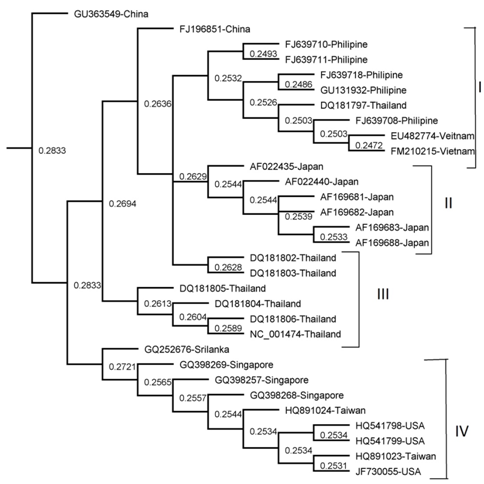
Figure 1 The phylogenetic tree was derived from NS3 sequence comparisons and constructed using the neighbour-
joining (NJ) method. The tree is shown with DEN-3 isolate, GU363549 as the out group. Significant bootstrap value is shown as percentages derived from 1,000 re samplings.
Tree made by UPGMA Tree
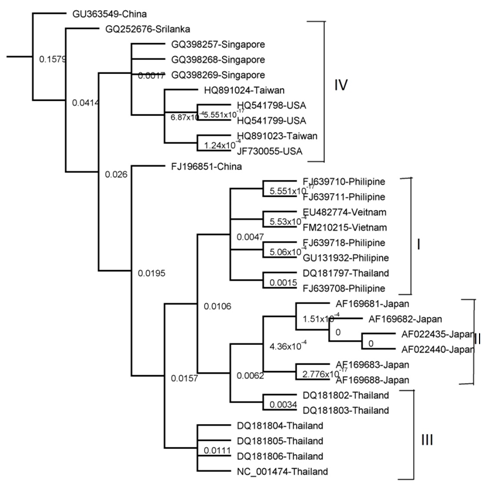
Figure 2 The phylogenetic tree was derived from NS3 sequence comparisons and constructed using the unweighted Pair Group Method with Arithmetic Mean (UPGMA) method. The tree is shown with DEN-3 isolate, GU363549 as the out group. Significant bootstrap value is shown as percentages derived from 1,000 re samplings.
Tree made by Mr. Bayes Method
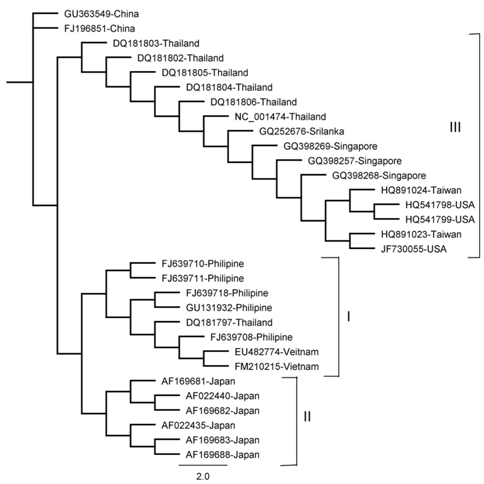
Figure 3 The phylogenetic tree was derived from NS3 sequence comparisons and constructed using the Mr. Bayes method. The tree is shown with DEN-3 isolate, GU363549 as the out group.
Tree made by Maximum Likelihood Method
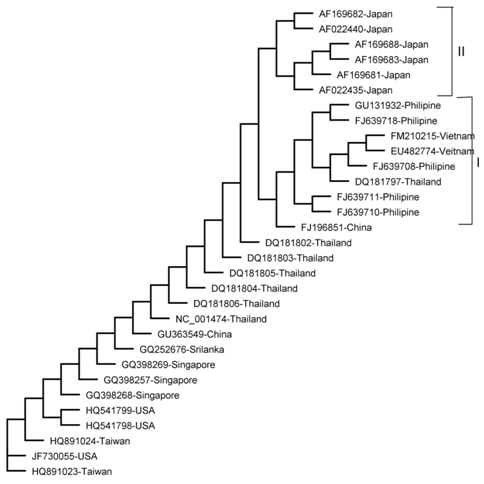
Figure 4 The phylogenetic tree was derived from NS3 sequence comparisons and constructed using the Maximum Likelihood (ML) method.
Tree made by Maximum Parsimony Method

Figure 5 A Maximum Parsimony tree depicting the phylogenetic relationships of the Dengue 2 isolates.
It is evident from our data that the Singapore, Sri Lanka, China, USA and Taiwan are the countries from where Dengue spread to other countries included in our study. It gives rise to the dengue 2 serotypes present in Thailand and from Thailand to Japan and Japan to Vietnam and Philippine.
Tree Topology
Tree made by NJ Method
A close similarity in the serotypes of Vietnam and Philippine was observed. They are found in the same clade in all the five phylogeny trees. Their clade showed a closest relation to the serotypes of Japan in our study. This shows that these serotypes have been derived from a common ancestor which is the serotypes of China and Thailand in our study.
Serotypes of Japan form a separate clade in all the tress. They show a close relation with Philippine, Thailand and China serotypes. Thailand serotypes were present in separate clade in NJ, UPGMA and Parsimony trees and in ML and Mr. Bayes tree these serotypes are showing close relation with the Japan, Philippine, Sri Lanka, Singapore and China. In this study we found that the Thailand serotype is the one which is closely related to most of the countries included in our study, so we can say that the serotypes of Thailand have played a significant role in the spread of dengue type 2 to the other countries of the world.
Serotypes of Sri Lanka, Singapore, USA and Taiwan are placed in the similar clades in our study in NJ, UPGMA and Parsimony trees and they share a clade along with serotypes of Thailand in Mr. Bayes tree. In ML tree they are not forming any clade but they are closely related to Thailand serotypes.
It is difficult to label one single ancestor of all other serotypes in our study because of intramethod variation. But it can be postulated that the serotypes of China, Sri Lanka, USA and Singapore all the likely serotypes which has given rise to dengue 2 serotypes present in Thailand. Thailand serotypes are present in an intermediate position in our study and they are interlinking both the ancestors with the descendants. The descendants of our study are the serotypes of Japan, Vietnam and Philippine.
In the present study we have found a closer similarity of NS3 sequence of dengue type 2 among Philippine and Vietnam. They form the same clade in all the methods employed. We have also found a close relation between the serotype of dengue 2 among Sri Lanka, China, Singapore, Taiwan and USA. Serotypes of Thailand and Japan form separate clades.
On the basis of our results we can predict that the serotypes of Singapore, Sri Lanka and China, USA and Taiwan are likely ancestors. Serotypes of Thailand are immediate descendants and serotypes of Japan, Philippine and Vietnam are descendant from serotypes of Thailand.
None.
None.

©2016 Atif, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.