Journal of
eISSN: 2373-6453


Review Article Volume 3 Issue 2
Centre for Infectious Diseases and Microbiology (CIDM), Institute of Clinical Pathology and Medical Research (ICPMR), Westmead Hospital, The University of Sydney, Australia
Correspondence: Fei Zhou, Centre for Infectious Diseases and Microbiology (CIDM), Institute of Clinical Pathology and Medical Research (ICPMR), Westmead Hospital, Darcy Road, Westmead, New South Wales, Australia, Tel (612) 9845 6255, Fax (612) 9893 8659
Received: February 16, 2016 | Published: February 26, 2016
Citation: Zhou F, O’Sullivan MVN, Iredell JR, Sintchenko V, Gilbert GL, Dwyer DE (2016) Molecular Analysis of Enteroviruses. J Hum Virol Retrovirol 3(2): 00083. DOI: 10.15406/jhvrv.2016.03.00083
Enteroviruses (EVs) are common human pathogens which usually infect the gastrointestinal and respiratory tracts and can spread to other organs or systems. They are characterized by high genomic plasticity, primarily due to high mutation and recombination rates. Improved molecular diagnostic methods and genetic sequence analyses are beginning to discover the complex characteristics of individual serotypes and genotypes. Understanding the tempo and pattern of molecular diversity and evolution is of great importance in the pathogenesis of EVs, information which will assist in disease prevention and control. In this review, we will focus on molecular analysis of EVs, including current diagnosis, epidemiology and evolution.
Keywords: Enterovirus, Molecular diagnosis, Molecular epidemiology, Molecular diversity, Evolution, EVs
PCR, Polymerase Chain Reaction; IRES, Internal Ribosome Entry Site; UTRs, Untranslated Regions; RT, Reverse Transcription; EVs, Enterovirus
The order Picornavirales consists of the families Picornaviridae, Dicistroviridae, Iflaviridae, Marnaviridae and Secoviridae and two unassigned genera, Bacillarnavirus and Labyrnavirus. The family Picornaviridae currently consists of 50 species grouped into 29 genera (http://www.picornaviridae.com). Enteroviruses (EVs) (genus Enterovirus, family Picornaviridae) are common human pathogens associated with a wide spectrum of conditions, ranging from asymptomatic infection to serious illness, such as aseptic meningitis, meningoencephalitis, myocarditis and acute flaccid paralysis, especially in children.1 and immunocompromised patients.2 The original classification of EVs consisted of polioviruses (PVs), coxsackieviruses A (CV-As) or B (CV-Bs) and echoviruses (Es), based on biological activity and disease. Currently, the genus EV is classified to 12 species, EV A to H, J and Rhinovirus
A to C (http://www.picornaviridae.com). Seven of the species, EV A to D (formerly
named Human enterovirusA to D) and Rhinovirus A to C (formerly named Human rhinovirus A to C) are known to infect humans.3
The EV genome is a positive single-stranded RNA molecule of approximately 7,500 nucleotides, comprising a single open reading frame flanked 5’ and 3’ by untranslated regions (UTRs) (Figure 1). The cloverleaf structure of domain (stem-loop) I in the 5’ UTR is important for viral replication;4 while domains II to VI encompass the internal ribosome entry site (IRES) which directs translation of the mRNA by internal ribosome binding.5 The coding region, divided into three subregions (P1, P2 and P3), consists of a single open reading frame encoding a polyprotein. The P1 region encodes four structural capsid proteins (VP4, VP2, VP3 and VP1). VP1, VP2 and VP3 are located at the surface of the viral capsid and are exposed to immune pressure, whereas VP4 is located inside the capsid.
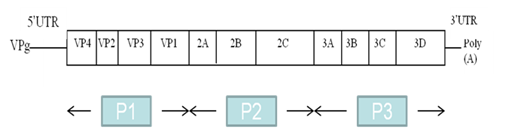
Figure 1 Structure of the enterovirus RNA genome, with the genome-linked protein VPg at the 5’ end, the 5’ UTR, the protein coding region (P1, P2 and P3), the 3’ UTR and the poly (A) tail. Coding regions for the viral proteins are indicated
The VP1 capsid protein is the most external and immunodominant of the picornavirus capsid proteins.6 and contains most neutralization epitopes. The P1 coding region for the capsid proteins provides reliable correlation between sequence relatedness and the previous definition of serotype.7 This also appears to be true for the various individual capsid protein regions, except for VP4, whose sequence does not always correlate with serotype.8,9 Recent molecular studies suggest that the VP1 nucleotide sequence correlates well with antigenic typing by neutralization and can be used for virus identification and evolutionary studies.7,10,11 VP1 sequencing has supported and clarified early serologic data, and assisted in proposals of new EV types.7,12,13 In addition, the non-structural proteins are encoded in the P2 (2A, 2B and 2C) and P3 (3A, 3B, 3C and 3D polymerase) regions.
Although the EV antigenic properties are very important, the introduction of molecular typing methods and a reassessment of the limitations of the original classification (PV, CV-A or CV-B and echoviruses) have resulted in the development of the current classification scheme. Currently, the genus EV is classified to 12 species, EV A to H, J and Rhinovirus A to C (http://www.picornaviridae.com). A picornavirus species is a class of phylogenetically related serotypes or strains which would normally be expected to share
Species |
(Sero) Types |
EV-A |
CV-A2 to -A8, -A10, -A12, -A14, -A16, EV-A71, 9 new types (EV-A76, -A89, -A90, -A91, -A92, -A114, -A119, -A120 and -A121) and the simian EVs SV19, SV43, SV46 and baboon EV A13 (BA13) |
EV-B |
CV-B1 to -B5 (including swine vesicular disease virus [SVDV]), -B6, -A9, 28 echovirus serotypes (E-1, -2, -3, -4, -5, -6, -7, -9, -11, -12, -13, -14, -15, -16, -17, -18, -19, -20, -21, -24, -25, -26, -27, -29, -30, -31, -32 and -33), EV-B69, 26 new types (EV-B73, -B74, -B75, -B77, -B78, -B79, -B80, -B81, -B82, -B83, -B84, -B85, -B86, -B87, -B88, -B93, -B97, -B98, -B100, -B101, -B106, -B107, -B110, -B111, -B112 and -B113) and the simian enterovirus SA5 |
EV-C |
PV-1 to -3, CV-A1, -A11, -A13, -A17, -A19, A20, -A21, -A22, -A24 and 11 new types (EV-C95, -C96, -C99, -C102, -C104, -C105, -C109, -C113, -C116, -C117 and -C118) |
EV-D |
EV-D68, -D70, -D94 and 2 new types (EV-D111 and -D120) |
Table 1 EV-A, B, C and D species and their corresponding (sero) types a
(http://www.picornaviridae.com).
In this review, we will focus on molecular analysis of EVs, including current diagnosis, epidemiology and evolution.
http://www.picornaviridae.com
Molecular diagnosis of enteroviral infections
The traditional approaches to detecting and characterizing EVs are based on the time-consuming and labour-intensive procedures of viral isolation in cell culture and neutralization with mixed hyperimmune equine serum pools and specific monovalent polyclonal antisera for confirmation.14 Due to distinct advantages in speed and convenience, molecular biology techniques are supplanting the traditional approaches of EV detection and characterization.10,14 Continued development and optimization of these methods will bring about further application of these techniques for EV diagnostics. As with virus isolation and serotyping, molecular approaches can detect the presence of EV in a specimen or confirm an isolate, and with some procedures, to further characterize the detected virus.15,16
The first category of nucleic acid tests is based on the polymerase chain reaction (PCR). This is applied primarily to detect EV genomes in clinical specimens, cell cultures and biopsy or autopsy tissues. The design of EV-specific primers (and probes) in the 5’ UTR to target all known EV members is probably practical and reasonable.14,16-19 Therefore, 5’ UTR EV-specific primers (and probes) can be used as an effective tool to confirm whether an isolate is an EV, and the design of generic assays lowers the risk of missing newly emerging or still unrecognized variants.20,21 Currently, PCR is commonly used to directly detect viruses in clinical specimens from individuals suspected of EV infection. The major advantage of the pan-EV PCR is that rapid EV detection is possible, even with very small amounts of clinical material such as cerebrospinal fluid.22 However, the sensitivity may be lower in certain specimens (e.g., stool).23 Furthermore, by changing the target, PCR can be used to characterize a particular EV. Certain genomic sequences within the capsid-coding region (especially VP1) correspond to the conserved antigenic property of the virus, since the antigenic property that defines serotype is a property of the viral capsid proteins.7 Investigations of the PV capsid regions have led to primer design that can selectively amplify isolates from a single serotype, but not from other heterologous serotypes.24 These principles have been extended to other EV serotypes or receptor groups, providing efficient methods for rapid EV characterization, such as type-specific reverse transcription (RT)-qPCR for sensitive detection of E30 in cerebrospinal fluid specimens.25 and a real-time RT-PCR assay for the detection of EV-D68 in clinical specimens.26
The second approach utilizes nucleic acid probes, and was the first application of molecular techniques to detect EV genomes. Early probes were made by synthesizing cDNA from CV-B viral RNA. Then, the cDNA was cloned into bacteria, amplified, and radiochemically labelled to provide a reagent for detecting EV genomes by nucleic acid hybridization.27 Further refinement of this technique included the use of RNA probes to improve sensitivity, the introduction of nonradiochemical probe labelling, and the application of these techniques to in situ hybridization in myocardial tissue.28,29 Application of bioinformatics in probe design enables detection of EVs from formalin-fixed tissue samples on different taxonomic levels by advanced in situ hybridization technology.30 Nucleic acid probe hybridization has also been broadly used in the WHO PV eradication program to determine whether an isolate is a vaccine-derived or a wild-type PV.31,32 Microarray techniques for the detection and characterization of EVs have been developed and applied in some studies, but they can only identify limited serotypes.33,34 In addition, a modified RT-PCR-based reverse line blot hybridization assay for synchronous detection of 5’ UTR and VP1 region has been developed as a quick, accurate and efficient approach to characterizing common and nonserotypeable EVs.11,14
The ease and low cost of this strategy will improve the diagnosis and epidemiologic investigation of EV infections and outbreaks where other typing methods are unavailable. The third group of molecular methods is based on EV genomic sequencing for characterization at the highest levels of specificity and accuracy.10,16,35 The information from the nucleotide sequence of a given virus represents its fundamental characterization.36,37 It is practical to use sequence information to assign an EV isolate to a particular (sero) type.37 The molecular typing system is based on RT-PCR and nucleotide sequencing of the 3’ half or the entire VP1 genomic region.11,35 and these can function as an excellent surrogate for serotyping by neutralization. The (sero)type of an unknown isolate is inferred by comparison of the partial or complete VP1 sequence with a database containing VP1 sequences for the prototype and variant strains of all known EV (sero)types. The principles of EV assignation include:
Other molecular methods that assist serotype-specific identification include combined multiplex RT-PCR and microarray to detect and differentiate EV-A71 and CV-A16.33 molecular characterization of PV strains by RT-PCR-restriction fragment length polymorphisms.40 an integrated micro-RT-PCR system for automatic detection of microorganisms including EV-A71.41 and an RT-loop-mediated isothermal amplification system for rapid and highly sensitive detection of EVs in stool samples.42
Molecular epidemiology of enteroviruses
Methods of molecular epidemiology: EVs can be transmitted by both faecal-oral and respiratory routes. Studies of the variations of enteroviral proteins or nucleic acids have provided significant epidemiologic information.43,44 Molecular epidemiologic investigations have aided in our understanding of EVs by:
Molecular typing approaches (including PCR and genomic sequencing) allow rapid and accurate identification of EVs. The major method applied to obtain epidemiologic information is direct analysis of genomic variations of multiple sequences (sometimes local sequences compared with GenBank database sequences) using different types of software (such as MEGA and BioNumerics) following nucleic acid sequencing in different regions of EVs.18,50 Phylogenetic analysis of sequences in different genes (such as 5’ UTR and VP1) can be used to discriminate between variants within a (sero) type, to enhance the epidemiologic linkage of EV strains, and to study the evolution of a given (sero) type or among different (sero) types.17-19 Analysis of whole genome sequences has been used to reveal epidemiologic and evolutionary dynamics of EVs through time and space.1,39,51 The combination of molecular typing and phylogenetic sequence analysis will assist with both individual patient diagnosis and public health measures. Moreover, some novel methods using bioinformatic technology have appeared. For example, the combination of multiple sequence alignment with the linear mapping hash function is a computationally efficient way of gene sequence clustering and can be a valuable tool for the assessment of similarity, clustering of different microbial genomes (such as VP1 region of EV-A71), identifying reference sequences, and for the study of evolution of bacteria and viruses.53
Molecular epidemiology of polioviruses
The application of nucleic acid sequencing to the studies of wild-type PV isolates from different areas has remarkably expanded the power of epidemiologic studies.53,54 Importantly, phylogenetic trees or lineage maps can be used for demonstration of sequence relationships.17,55,57 Closely related viruses can be easily detected by analysing random mutations in different PV genomes; while more distantly related viruses can be clustered into distinct geographic groups of endemic circulation. From 2001, the WHO polio network laboratories routinely sequence the complete VP1 gene of all wild-type and vaccine-derived PV isolates for comparison. Using VP1 sequence analysis, the genetic diversity of PV strains can be exploited in molecular surveillance.55
Moreover, the rapid evolution of PVs permits the use of comparative VP1 nucleotide sequencing to resolve the fine structure of a poliomyelitis outbreak, and to elucidate individual chains of transmission.58 Based on a nucleic acid sequence database of PV strains worldwide, it has been possible to develop rapid approaches to tracking wild-type PV strains.59 Recently, in response to the observation that the large majority of vaccine-derived PV isolates are type 2, WHO has developed a plan for coordinated worldwide replacement of trivalent oral PV vaccines (tOPV) with bivalent OPV (bOPV; types 1 and 3) in April 2016, preceded by the introduction of at least one dose of inactivated PV vaccine into routine immunization schedules worldwide.60
Molecular epidemiology of non-polio enteroviruses
Research on the molecular epidemiology of non-polio EVs has concentrated on the evolutionary features following the comparison of viral isolates within a (sero)type over time.19,61 the comparison of isolates from different (sero)types.18,62 and between different genera within the Picornaviridae.63
EV-A71 and CV-A16 are members of EV-A species, and are major and independent etiologic agents of hand-foot-mouth disease .64,65 To understand the molecular epidemiology and evolution of EV-A71, whole-genome sequences.1 and several distinct regions of the EV-A71 genome have been employed for analysis, such as 5’ UTR.19,66 3D polymerase region.67 VP4 region.68 and VP1 region.19,49,61,69,70 However, the VP1 gene is considered to be the most informative and robust region for evolutionary study due to a high degree of diversity and lack of involvement in recombination.71 EV-A71 strains isolated worldwide are classified into four genogroups: A-D.72
Genogroups B and C have been differentiated into subgenogroups B0-B5 and C1-C5.72 Recent studies have proposed three new genogroups, including E (in sub-Saharan Africa), F (in Madagascar).73 and G (in India).74 Other new subgenogroups proposed recently include C4a, C4b, C6, C7 and B6.75,76 The analysis of EV-A71 based on VP1 region, in combination with 5’ UTR.19 or 3D polymerase region.67 could provide more objective information on molecular epidemiology and evolution of EV-A71. For CV-A16, the VP1 and VP4 genes are mostly used for classifying the virus.19,77-79 It has been classified on VP1 analysis into geno groups A and B, with geno group B further divided into subgenogroups B1 (including B1a, B1b and B1c), B2.64,80 and a possible B3.19 Molecular analysis of VP1, combined with 5’ UTR, may also enhance the reliability of molecular surveillance and the understanding of genetic evolution of CVA16.19
In EV-B species, several CV-B outbreaks or epidemics have been recorded. CV-B1 infection accounted for 23% of reported cases with severe neonatal morbidity and multiple deaths between 2007 and 2008 in the USA.81 CV-B5 has caused widespread epidemics in the USA, particularly in 1967, 1972 and 1983.82 The sequence analyses of these CV-B viruses have been based on VP1 region, 5’ UTR and 3D.18,81-84 Four main genotypes (GI-GIV) and four small clusters (C1-C4) of CV-B1 have been described based on the VP1 region.84 E11 causes a full spectrum of enteroviral diseases, including meningitis,85 but it has been particularly associated with cases of severe neonatal disease.86 A few studies have analysed the genetic diversity among E11 VP1 (-2A), 3D polymerase and 5’ UTR sequences,18,85-87 which divided E11 strains into different subgroups. The E11-B cluster has been shown to represent a phylogenetically distinct subgroup (with altered virulence) within the E11 serotype.87 Since its characterization in 1958, E30 has been increasingly associated with meningitis outbreaks worldwide.88 Several studies have shown different clustering of E30 variants based on VP1 and/or 5’ UTR sequences.18,89-91
Investigations have shown that EV-C (sero) types (including PVs) cluster into three subgroups in the VP1 region, designated as A to C.92,93 CV-A11 groups together with the strains of subgroups B or C depending on the method of phylogenetic tree construction.93 The major worldwide CV-A24 epidemics have been attributed to four genotypes of CV-A24 variant strains (I to IV) on the basis of phylogenetic analysis of the 3C protease or VP1 region.94,95
After the initial identification of EV-D68 (belonging to EV-D species) in 1962, detection of this virus was rarely reported until the early 2000s. EV-D68 has caused widespread respiratory illness recently. Neurological syndromes including acute flaccid paralysis following EV-D68 infection have also been reported in a small number of cases.39,96 Whole-genome sequences39 and several distinct regions of the EV-D68 genome have been used for analysis of molecular epidemiology, such as 5’ UTR, VP1 and VP4/ VP2 regions.97,98 The EV-D68 strains circulating in recent years are classified into three genetic groups.99,100 For the VP1 region, strains belonging to lineages 1 and 2 are likely to form clusters containing viruses from the same geographical origins on the phylogenetic tree. In contrast, strains belonging to lineage 3 are less likely to show geographical clustering patterns. These data suggest that transmission at the global level might be more common for lineage 3 than for lineages 1 and 2.101
Molecular diversity and evolution of enteroviruses
Most RNA virus genomes have much higher evolution rates than DNA viruses.102 EVs are well known for high genomic plasticity due to high mutation and recombination rates. Analysis of whole genome sequences has been used to reveal epidemiologic and evolutionary dynamics of EVs through time and space.1,39,51 Studies of EVs suggested that sequence variations in the UTRs, the VP1 region and the 3D polymerase could affect virus infection and replication capability in vitro and in vivo.103,104
Mutation of enteroviruses
Base misincorporation during chain elongation and lack of 3’ to 5’ exonuclease proofreading ability in RNA polymerases can cause very high error rates in EVs; there is a rate of spontaneous mutation of approximately one mutation per genome per replication.105 High RNA virus error rates are necessary to enable survival of the virus population under selective pressure.106 The molecular diversity and evolution of EVs occurs through genetic drift and, over much longer periods, antigenic diversification in the structural gene region encoding the virus capsid, including VP1.61,107 Sequence changes occur rapidly in the capsid region.61 Studies have also proved that mutations in the 5’ UTR markedly decrease multiplication efficiency,108 alter cell tropism,109 and attenuate virulence.110
In EV-A species, the overall rapid evolutionary rate of the EV-A71 VP1 gene has been estimated to be 4.2 × 103 and 3.4 × 10-361 or 4.5 -4.6 × 10-3 and 4.2 × 10-349 all substitutions per nucleotide per year for genogroups B and C, respectively. The Ala to Val substitution at VP1 position 170 appears to be associated with increased neurovirulence.69 Additionally, the nucleotide mutation (A→G) at position 485 in the 5’ UTR may be one of the molecular determinants of EV-A71 attenuation of neurovirulence in cynomolgus monkeys.111 The average evolutionary rate calculated for CV-A16 B1a and B2 was 0.91 × 10-2 synonymous substitutions per nucleotide per year based on VP1 gene.64 The evolutionary rate of CV-A16 is relatively slow given that the prototype CV-A16 strain was first identified in South Africa in 1951, and that all other CV-A16 strains formed a single genogroup B following approximately 60 years of evolution.64 Comparing two regions (5’ UTR and VP1) of EV-A71 and CV-A16 viruses provides evidence of epidemiologic linkage of EV-A strains, and mutation in the two regions plays a vital role in the evolution of these viruses.19
In EV-B species, the broad spectrum of CV-B associated disease reflects the existence of multiple strains within a single serotype with various degrees of virulence.112 Individual nucleotide substitutions in the noncoding and coding regions of the viral genome determine virulence.113 Major pathogenic determinants of CV infections have been localized to the 5’ UTR and the capsid protein coding region.114 The estimated rate of CV-B1 evolution in the VP1 gene was 7.73 × 10-3 substitutions per site per year.84 Strong negative selection shaped the evolution of VP1 and 3CD loci in CVB5 strains, but compelling phylogenetic data suggested that immune selection pressure resulted in the emergence of the two genogroups (A and B) with opposed evolutionary pathways.115 The genogroups A and B also differed in the temporal occurrence of the amino acid changes. Studies of E30 meningitis outbreaks have shown that point mutations give rise to substantial genetic diversity in VP1. Substitutions in VP1 region occurred predominantly at synonymous sites, with VP1 showing a rapid substitution rate of 8.3/8.4 × 10-3 substitutions per site per year, and recombination frequency was tightly correlated with VP1 divergence.116,117 The predicted rates of evolution in the VP1 region for E9 and E11 were 5.8 × 10-3 and 4.8 × 10-3 substitutions per site per year, respectively.118 Moreover, the study of Spanish E6 strains revealed high rates of sequence change of the VP1 region (around 1.1× 10-2 substitutions per site per year) and there was a direct relationship between recombination frequency and VP1 sequence divergence for E6.119 The relatively high sequence diversity among the 5’ UTR and VP1 regions of serotypes in EV-B species suggests that nucleotide substitution is probably the dominant evolutionary mechanism in these regions of the genome.18
In EV-C species, PVs are among the most rapidly evolving viruses known, with most evolution appearing to be random genetic drift since >80% of nucleotide substitutions within the coding region generate synonymous codons. Nucleotide substitutions accumulate at an overall rate of approximately 1% per year at all sites, and at approximately 3% per year at synonymous sites, calculated primarily on the VP1 region or P1/capsid region.106,120,121 Evolution rates appear to be similar across serotypes and between wild-type and vaccine-derived PVs.56,106 A common feature of the Sabin oral PV vaccine strains is the presence of nucleotide substitutions in the IRES of 5’ UTR, which in serotypes 1 and 3 are critical attenuating mutations. Additional mutations encoding amino acid substitutions in the capsid region (including VP1) contribute to and stabilize the attenuated phenotype.122 In CV-A24 variant strains, the estimated evolutionary rates were 1.0 - 4.1 × 10-3 or 6.27 - 6.67 × 10-4 per nucleotide per year based on the 3C protease gene,94,123-125 4.2 × 10-3 per nucleotide per year based on the VP4 gene,123 and 1.83-2.17 × 10-3 per nucleotide per year based on the 3’ VP1 gene.125
The mean nucleotide substitution rates of EV-C subgroup B (ranging from 1.170 to 3.625 × 10-3 with combined high-probability distribution interval of 1.170 to 3.625 for partial VP1) are slightly lower than the corresponding estimates for other EV serotypes.93 Intraserotypic genetic change of VP1 is dominated by silent mutations accompanied by amino acid polymorphism occurring dominantly at immunogenic sites, while interserotypic differences of VP1 include permanent fixation and insertion/deletion of distinct ‘signature’-amino acids that could be a result of larger scale changes in the capsid structure.93 The relatively high sequence diversity among the 5’ UTR and VP1 regions of EV-C viruses also suggests that nucleotide substitution is probably the dominant evolutionary mechanism in these regions of the genome.17
In EV-D species, the mean rate of nucleotide substitution for VP1 gene of EV-D68 was 6.2 × 10-3 substitutions per site per year.97 Mutations have accumulated in two specific regions of the EV-D68 VP1 sequences: the BC and DE loops.100 Selection analyses conducted on the VP1 amino acid sequences in several studies have revealed that most of the positively selected codons are found in the BC and DE loop regions.126,127 These findings suggest that antigenic epitopes might be located in the BC and DE loop regions. Unique amino acid mutations in the VP2 and VP3 regions that can differentiate recently detected strains from the Fermon strain, as well as recent strains of one genetic lineage from another, have been identified; however, such mutations were found less frequently than those in the VP1 region.127 Strains of lineage 1 were likely to have deletions at both positions 681-704 and 721-731 in the 5′UTR. In contrast, lineage 3 strains had deletions only at positions 624-704, whereas those of lineage 2 had no deletions in the 5′UTR.101
Recombination of enteroviruses
The evolutionary diversity of EVs is also due to intraserotypic / interserotypic (or interspecies) recombination, the majority of which occurs within P2, P3 non-structural coding regions and 5’ UTR, with little or no recombination within the P1 capsid coding region;85,128-130 however, the survival of offspring with recombination within the P1 capsid coding region is rare.8,128 In general, only the P1 capsid region (mainly VP2-VP3-VP1) is inherited as a single unit, suggesting that it is the primary determinant of EV identity.129-131 Due to recombination, no general correlation is found between sequence relatedness and specific serotype in genome segments outside the capsid region.
RNA recombination is believed to be coupled with the process of genome RNA replication. It occurs by template switching during negative strand synthesis, as first demonstrated in PV-infected cells132 and subsequently in cell-free extracts.133 Recombination can occur between PVs and other EV-C viruses within the 5’ UTR and non-structural coding regions; this shuffling of different non-structural coding regions may lead to serotypes with selective advantages.3,46,120 Recombination within the 5’ UTR and the non-structural coding regions was also found among EV-A and EV-B members (within the same species).129,134 The frequency of recombination differs between EV species, being higher in EV-B species than in EV-A species8 and very low in EV-D species.135,136
In EV-A species, EV-A71 intraserotypic and interserotypic recombination have been reported.19,67,137 Recombination would be facilitated by the presence of several subgenogroups circulating during the same period in one country or region, such as the co-circulation of four subgenogroups B3, B4, C1 and C2 in Malaysia between 1997 and 2000.138 Recent studies revealed that some CV-A16 isolates might have recombination with the EV-A71 isolates in the 5’ UTR or 2A-2B region, and such recombination events might change the epidemic trend.80,139
In EV-B species, although the major neurotropism determinant in the CV-B3 genome was in the capsid region, viruses containing the CV-A9 capsid were also able to initiate infection in the central nervous system provided they contained the CV-B3 5’ UTR.114 The presence of the 5’ UTR of CV-A9 clearly enhanced muscle tissue tropism. The viability of chimeric viruses constructed between CV-A9 and CV-B3 suggested that recombination could take place in nature, partly explaining the genetic diversity and pathogenic variation among CVs.114 Studies have demonstrated that recombination can occur between the 5’ UTR and capsid, the capsid and 3D, or both in some CV-B cases.18,84,140
Recombination could potentially lead to new strains that might cause outbreaks of disease. Furthermore, studies of echoviruses suggest that recombination has played a significant role in the evolution of these echoviruses. For example, E30 can recombine with any EV-B strain in the non-structural region or 5’ UTR.18,90,141 In the non-structural coding region of E9, E11 and E30, sequence change has been shown by frequent recombination events, leading to the acquisition of novel 3D polymerase sequences interspersed with sequences of other EV-B isolates.118 The first evidence for natural intertypic recombination for E24 was described recently, which showed that this E24 strain recombined with a CV-B1 stain at the 2C region.51 In fact, interserotypic recombination may play a major role in the evolution of some EV-B viruses.18 Due to the need of gene regulation, restricted genetic variation has been noted mainly in the non-structural coding region, and in some cases recombination occurs in the 5’ UTR.
In EV-C species, natural interserotypic recombination within the capsid region of PVs appears to be rare, probably because structural incompatibilities restrict the fitness of most interserotypic recombinants.142 In contrast, interserotypic recombinants excreted by recent vaccinees exposed to tOPV and recombinants among circulating wild-type PVs and vaccine-derived PVs usually have crossovers restricted to the noncapsid region.142 PVs recombine actively with each other and with other closely related EV-C members, a process which might offset the effects of accumulation of deleterious mutations arising in some lineages.46,121,142,143 In fact, the recombination between PVs and other EV-C viruses is a normal feature of PV evolution in nature. Recombination may lead to a lack of linkage between noncapsid sequences and the neutralisation serotype of a virus within a species.46
In addition, the prototype EV-C strains are recombinant relative to each other,92 and circulating EVs commonly recombine with wild-type and vaccine PVs.144 Studies have shown that, compared with VP1, the 5’ UTR of CV-A (sero)types and PVs has frequently been subject to recombination,17,145 indicating that interserotypic recombination may play a major role in the evolution of some of these viruses. The phylogenetic pattern of the EV-C strains in a recent study suggests non-random recombination between strains of the same species.3 The frequency of recombination appears to differ between species, types and intraspecies groupings, which is possibly due to cell tropism, sequence similarity and the phylogenetic position of a given virus strain.3
Recombination also occurs in new EV (sero) types. For instance, the 5’ UTR of an EV-C109 strain in a study is likely the product of an interspecies recombination event between ancestral members of the EV-A and EV-C groups.146 Another analysis of the P3 region of an EV-C104 strain indicates that this virus has undergone a rearrangement within the last decade, likely due to recombination with an EV-C117-like virus.130
Analyses of EV prototype strains and clinical isolates suggest that interserotypic recombination is a frequent event and that it generally occurs among viruses of the same species. The exception is in the 5’ UTR, where only a single genetic group can be identified within EV-A and -B species and a second within EV-C and-D species.129,134,137 This reflects a different pressure for evolutionary change exerted on the 5’ UTR and the rest of the genome, largely shaped by the functional significance of the specific genomic region and its interaction with the stable cellular environment.147,148 Likewise, recombination may delink a given capsid sequence (serotype) from its original 5’ UTR, explaining why 5’ UTR is generally unsuitable as a molecular alternative to serotype identification.129 In general, the EV genome could be recognized as a stable symbiosis of genes, and EV species consist of a finite set of capsid genes responsible for different serotypes and a continuum of non-structural protein genes that seem to evolve in a relatively independent manner.149
EVs are characterized by high genomic plasticity, primarily due to high mutation and recombination rates. Improved molecular diagnostic methods and genetic sequence analyses are beginning to discover the complex characteristics of individual serotypes and genotypes. Understanding the tempo and pattern of molecular diversity and evolution is of great importance in the pathogenesis of EVs, information which will assist in disease prevention and control.18
The vast majority of EVs infect the gastrointestinal and respiratory tracts, and can spread to other organs or systems. Improvements in surveillance, serological surveys and detailed genetic and antigenic characterization of viral populations may help to elucidate biological mechanisms.150 Better characterization of these human pathogens may help to develop vaccines or antiviral treatments and to monitor the emergence of new strains.151
None.
None.

©2016 Zhou, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.