Journal of
eISSN: 2373-6453


Research Article Volume 2 Issue 2
Centre for Infectious Diseases and Microbiology (CIDM),
Institute of Clinical Pathology and Medical Research, Australia
Correspondence: Fei Zhou, Centre for Infectious Diseases and Microbiology (CIDM), Institute of Clinical Pathology and Medical Research (ICPMR), Westmead Hospital, Darcy Road, Westmead, New South Wales, Australia, Tel (612) 9845 6255, Fax (612) 9893 8659
Received: January 13, 2015 | Published: March 16, 2015
Citation: Zhou F, O’Sullivan MVN, Iredell JR, Sintchenko V, Gilbert GL, et al. (2015) Molecular Analysis of Enterovirus C Species Using the 5’ Untranslated Region and VP1 Region. J Hum Virol Retrovirol 2(2): 00038. DOI: 10.15406/jhvrv.2015.02.00038
Enteroviruses (EVs) of EV-C species evolve quickly, mainly due to high mutation and recombination rates. The 5’ untranslated region (UTR) is fundamentally important for efficient viral replication and for virulence; the VP1 region correlates well with antigenic typing by neutralization, and can be used for virus identification and evolutionary studies. In order to investigate the evolutionary diversity and molecular epidemiology in EV-C species, the 5’ UTR and VP1 regions were analyzed in 16 clinical isolates from a single public-health laboratory (serving New South Wales, Australia), representing seven types (serotypes/genotypes) in EV-C. Sequences were compared with the 5’ UTR and VP1 regions of 183 strains available in GenBank, representing the same seven types. The genetic relationships were analyzed using MEGA. The sequence analyses of the 5’ UTR and VP1 regions of 199 EV-C strains demonstrated that:
Keywords: Enterovirus C, 5’ UTR, VP1 gene, Molecular diversity, Molecular epidemiology.
EVs, Enteroviruses; UTRs, Untranslated Regions; IRES, Internal Ribosome Entry Site; PV, Poliovirus; RT-PCR, Reverse Transcription-PCR; VDPVs, Vaccine-Derived PVs; MAD, Madagascar;
Enteroviruses (EVs) (genus Enterovirus, family Picornaviridae) are small RNA viruses associated with a variety of human diseases ranging from asymptomatic or mild infections to more severe diseases such as aseptic meningitis and acute flaccid paralysis.1 The EV genome is a positive single-stranded RNA molecule of approximately 7,500 nucleotides, comprising a single open reading frame flanked 5’ and 3’ by untranslated regions (UTRs). In the 5’ UTR, the cloverleaf structure of domain (stem-loop) I is important for viral replication2,3 while domains II to VI encompass the internal ribosome entry site (IRES) which directs translation of the mRNA by internal ribosome binding.4 The 5’ UTR is relatively conserved at the nucleotide level, and is often used for nucleic acid testing to diagnose EV infections.5,6
The coding region, divided into three sub regions (P1, P2 and P3), encompasses a single open reading frame encoding a poly protein. The P1 region encodes four structural proteins (VP4, VP2, VP3 and VP1); the non-structural proteins are encoded in the P2 (2A, 2B and 2C) and P3 (3A, 3B, 3C and 3D polymerase) regions. VP1, VP2 and VP3 are located at the surface of the viral capsid and are exposed to immune pressure, whereas VP4 is internal.7 The VP1 capsid protein is the most external and immuno dominant of the picornavirus capsid proteins8 and contains most neutralization epitopes.
Currently, EVs are classified to twelve species, Enterovirus A to H, J and Rhinovirus A to C (http://www.picornaviridae.com). Seven of the species, Enterovirus A to D (formerly named Human Enterovirus A to D) and Rhinovirus A to C (formerly named Human rhinovirus A to C) are known to infect humans.9 EV-C species comprises three poliovirus (PV) serotypes (PV1, -2 and -3), nine coxsackie virus A (CVA) serotypes (CVA1, -11, -13, -17, -19, -20, -21, -22 and -24) and 11 new types. VP1 sequences correlate well with antigenic typing by neutralization, and can be used for virus identification and evolutionary studies.10,11 Molecular typing methods largely depend on reverse transcription-PCR (RT-PCR) amplification and nucleotide sequencing of the entire or 3’ half of the VP1 gene.1 Comparison of individual VP1 sequence with databases of VP1 sequences of EV prototype and variant strains allows genotype assignation and identification of new EVs.12
Four EV-C serotypes (including three PV serotypes and CVA24) have been identified from sero typed isolates in a single laboratory (serving New South Wales, Australia) from 1979 to 2007. In addition, 3.6% (227/6383) of EVs were non sero type able by neutralization. Three EV-C genotypes (CVA11, -20 and -21) were identified from non sero type able EV isolates by partial VP1 sequencing.13 Therefore, a total of seven EV-C types (serotypes/genotypes) have been identified in our laboratory over 29 years. In order to investigate evolutionary diversity and molecular epidemiology in EV–C, the nucleotide sequences of 5’ UTR and VP1 regions of 16 clinical isolates (representing these seven EV-C types) were compared with those of the 5’ UTR and VP1 regions of 183 strains (representing the same seven types) available in GenBank.
Viruses: The laboratory database contains 6383 EVs (‘local isolates’) between 1979 and 2007 from clinical samples. Isolates were obtained by inoculation of clinical specimens, including specimens in which EV had been detected by real-time fluorescence PCR, into cultures of monkey kidney cells (primary, secondary or cell lines) and various continuous cell lines, including A549 (human lung carcinoma), MRC-5 (human embryonic lung fibroblast), and RD (human rhabdomyosarcoma) cells. Prior to 2002, the titer of each cell culture with Enterovirus cytopathic effect (CPE) was first determined, and then a neutralization assay was performed in tissue culture tubes by using intersecting pools of hyper immune horse sera (National Institute of Allergy and Infectious Diseases, Bethesda, Maryland, USA) against 100 50% tissue culture infective doses (TCID50) of each isolate. If necessary, further identification was performed using a monovalent horse antiserum corresponding to the serotype identified by the intersecting pools. After 2002, group specific monoclonal pools and/or type specific monoclonal antibodies (Chemi-Con, Temecula, CA, USA) were used as a rapid identification alternative and neutralization was performed on isolates that were not identified. EV isolates that were not identifiable by standard sero neutralization procedures were labeled as ‘nonserotypeable’ isolates. Virus isolates were stored as unpurified cell culture supernatants at -70°C after initial isolation and sero typing.13,14 Beyond this they were usually not further passages. This study analyzed 16 EV-C local isolates representing seven EV-C types, (Table 1), including five initially classified as nonserotypeable by neutralization.
Serotype/genotype |
No. of sequences from GenBank (prototype strain GenBank no.)a |
Laboratory identifierb |
Age (months-M or years-Y) |
Specimen type |
5’ UTR GenBank accession number of local isolate |
VP1 GenBank accession number of local isolate |
PV1 |
69 (wild prototype strains [V01149 and AY560657], Sabin 1 vaccine strain [V01150 or AY184219]) |
03.295.3171 |
3M |
Nasopharyngeal aspirate |
GU236309 |
- |
05.354.2186 |
5M |
Nasopharyngeal aspirate |
GU236310 |
- |
||
03.309.2667 |
6M |
Nasopharyngeal aspirate |
- |
- |
||
PV2 |
60 (wild prototype strain [M12197], Sabin 2 vaccine strain [X00595 or AY184220]) |
102373.91 |
5M |
Stool |
GU236311 |
- |
90181.91 |
3M |
Stool |
- |
FJ868310 |
||
21554.91 |
4M |
Stool |
- |
- |
||
PV3 |
13 (wild prototype strain [K01392], Sabin 3 vaccine strain [X00925, AY184221 or X00596]) |
50660.91 |
6M |
Stool |
GU236312 |
FJ868312 |
82635.91 |
4M |
Stool |
GU236313 |
- |
||
106210.91 |
6M |
Stool |
GU236314 |
FJ868311 |
||
9245.91 |
5M |
Stool |
- |
FJ868336 |
||
CVA11 |
3 (AF499636) |
03.105.2708# |
23Y |
Stool |
GU236304 |
GU142908 |
CVA20 |
12 (AF499642) |
00.193.2216# |
1Y |
Stool |
GU236305 |
GU142909 |
|
38672.91# |
2Y |
Swab |
GU236306 |
GU142910 |
|
CVA21 |
14 (AF546702) |
98.072.1407# |
6Y |
Swab |
GU236307 |
GU142911 |
CVA24 |
12 (EF026081) |
06.278.4269.1# |
34Y |
Eye |
GU236308 |
FJ868371 |
|
06.278.4269.2 |
34Y |
Eye |
- |
- |
Table 1 The 16 EV-C local isolates used for analysis of 5’ UTR and VP1 regions
Note:
PCR primers and sequencing primers for 5’ UTR and VP1:
published primers for PCR amplification (5UTR-S and 5UTR-A1) and sequencing (5UTR-S and 5UTR-A2) of partial 5’ UTR were used.15 Published primers 494 to 497 for PCR amplification and sequencing of complete VP1 were obtained,16 as well as PCR amplification and sequencing primers 292 and 222 for analysis of partial VP1 sequences.16 Primer sequences were evaluated using the Sigma DNA Calculator (Sigma, http://www.sigma-genosys.com/calc/dnacalc.asp), and synthesized by Sigma-Aldrich (Sydney, NSW, Australia).
RT-PCR for 5’ UTR and VP1:
Viral RNA was extracted from 200 µl of virus-infected cell culture supernatant by use of a High Pure viral RNA kit (Roche Applied Science, Mannheim, Germany) according to the manufacturer’s procedures and eluted with 50 µl elution buffer. Reverse transcription (RT) was performed as described previously.17 EV-specific 5’ UTR PCR (using primers 5UTR-S and 5UTR-A1) for all 16 EV local isolates and partial VP1 PCR (using primers 292 and 222) for four isolates were performed as described previously.15 The methods of complete VP1 PCR for 12 EV-C isolates were as described previously.17
Sequencing and analysis for 5’ UTR and VP1:
Sequencing methods were described previously.17 For 5’ UTR sequencing of all 16 local isolates, primers 5UTR-S and 5UTR-A2 were used; for sequencing partial VP1 of four isolates, primers 292 and 222 were employed.15 The complete VP1 gene for each of 12 EV-C isolates was sequenced in two fragments using primers 494 to 497, followed by assembly of the sequences of the two fragments. The previously tested serotype for each of 11 sero typed isolates or the genotype for each of five nonserotypeable isolates (Table 1) was confirmed by pair wise comparison of the complete or partial VP1 sequence with a database containing VP1 sequences for the prototype and variant strains of all known EV serotypes.12
Comparison with GenBank database sequences. For molecular analysis, the 5’ UTR and VP1 regions of the 16 local isolates were compared with the 5’ UTR and VP1 regions of 183 strains (Table 1) available in GenBank representing the same seven types. The length for each of the 183 GenBank sequences was at least 3,200 nucleotides, including both 5’ UTR and VP1 regions. Sequences were trimmed and downloaded separately into two databases for 5’ UTR and VP1 using Bio manager.17 Most of the 183 GenBank sequences represent strains isolated from disease outbreaks from different locations over time; a small number of sequences were generated from asymptomatic or healthy people.18 Several strains were isolated from the same patient over time (e.g., in a study of long-term evolution of Sabin PV1 in an immuno deficient patient,19 and a small number were laboratory-modified strains.20 Where available, further information (including isolation dates and countries of origin) that could not be found directly from GenBank was obtained from the associated references.
Data analysis for 5’ UTR and VP1:
Two groups (PVs; CVA11, -20, -21, -24 and PVs) were analyzed separately. Alignment of all 5’ UTR and VP1 nucleotide sequences was undertaken intra-typically and inter-typically for EVs from EV-C using the ClustalW (accurate) program of Bio manager. Phylogenetic trees of 5’ UTR and VP1 regions were reconstructed by neighbor joining using the nucleotide/Kimura two-parameter method with MEGA, version 521 (Figures 1 & 2). Bootstrap analysis with 1,000 pseudo replicates provides an estimate of reliability for phylogenetic reconstructions. The bootstrap values in 1,000 pseudo replicates within trees are shown as percentages.
Nucleotide sequence accession numbers:
The GenBank accession numbers for the 5’ UTR sequences of the local isolates are GU236304 to GU236314; and for the VP1 sequences are FJ868310 to FJ868312, FJ868336, FJ868371, and GU142908 to GU142911 (Table 1).
Sequencing results for 5’ UTR and VP1
The length of the 5’ UTR sequences for the 16 local isolates ranged between 487 and 493 nucleotides, covering the relatively conserved major part of the IRES. The relatively long 5’ UTR sequences amplified by one consensus primer pair (5UTR-S and 5UTR-A1) provided a more accurate and systematic interpretation for 5’ UTR.15
The deduced amino acid sequences from the partial or complete VP1 sequences included the B-C loop, a region needed for type-specific antibody reactivity.22 The VP1 genotypes of all 11 sero typed isolates corresponded with their respective serotypes; for the five nonserotypeable isolates, genotypes were determined by VP1 sequencing. All seven types (serotypes/genotypes) analyzed in this study are listed in (Table 1).
Background of 16 local EV-C isolates
Ages of the patients (or asymptomatic PV Sabin vaccine recipients) and specimen types of the 16 local EV-C isolates are presented in (Table 1). These EV-C isolates were cultured from stool, nasopharyngeal aspirates, eye or other swabs.
Data analysis for PV1, -2 and -3
The phylogenetic relationships among 72 PV1 strains, 63 PV2 strains and 17 PV3 strains, including 10 local isolates (Table 1) isolated from asymptomatic Sabin vaccine recipients, were inferred by the neighbor-joining method using the nucleotide/Kimura two-parameter method with MEGA, based on alignment of the complete VP1 and partial 5’ UTR nucleotide sequences (Figure 1).
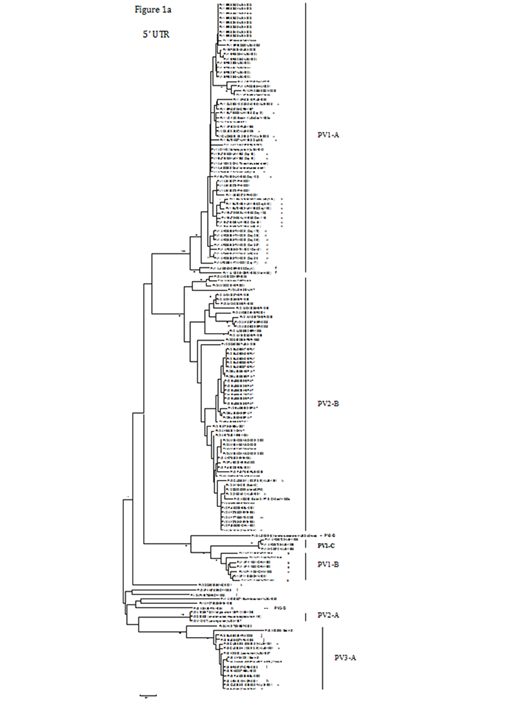
Figure 1A Phylogenetic trees showing the genetic relationships among 72 PV1 strains, 63 PV2 strains and 17 PV3 strains (including 10 local isolates, [labelled ‘a’, ‘b’ and ‘c’, respectively], with laboratory identifiers provided in parentheses) based on alignment of the complete VP1 (906, 903 and 900 nucleotides, respectively) and partial 5’ UTR (489, 490 and 492 nucleotides, respectively) sequences. Trees were constructed by neighbour joining using Nucleotide/Kimura 2-parameter method with MEGA, version 5.0.21 Bootstrap values (percentage of 1,000 pseudo replicate data sets) of ≥75% supporting each cluster are shown at the nodes. The scale bars represent the genetic distance.
Three primary groups for PV1, -2 and -3 were evident from the VP1 phylogenetic tree (Figure 1b). The PV1 group could be classified into three subgroups, PV1-A, -B and -C. The largest subgroup was PV1-A, which contained two wild prototype PV1 strains (V01149 and AY560657), the Sabin 1 vaccine strain (V01150 or AY184219) and other PV1 strains isolated from different countries or areas. Three local strains (labelled ‘a’) isolated from 2003 to 2005, with high nucleotide identity (ranging from 99.4 to 100%) to the Sabin 1 strain (V01150), were included in PV1-A. Several poliomyelitis outbreaks associated with type 1 circulating vaccine-derived PVs (VDPVs) have been reported in the Philippines in 200123 and in Hispaniola from 2000 to 2001.24 Three special examples are interesting. Nine isolates (labelled‘d’) from an immuno deficient Taiwanese patient over a 337-day period after onset of paralysis25 differed from Sabin 1 by 2.4 to 3.5% in VP1. In another case, 12 strains (labelled ‘e’) isolated over a 649-day period from a hypogamma globulinemic patient after oral vaccination with Sabin 1 in the United Kingdom19 showed high nucleotide identity (98 to 100%) to the Sabin 1 strain. In the third example, PV1 was isolated from an immuno deficient patient four days after onset of paresis and 66 months (5.5 years) of prolonged enteral virus replication.26 These two isolates (labelled ‘f’) differed from Sabin 1 by 5.3% and 9.2%, respectively. The second subgroup, PV1-B, comprised seven strains (including five strains labelled ‘g’) isolated on the Chinese mainland from 1989 to 1993,27 with ≥ 93.5% nucleotide identity. The third subgroup was PV1-C, which only comprised three strains from Albania during the 1996 outbreak.28
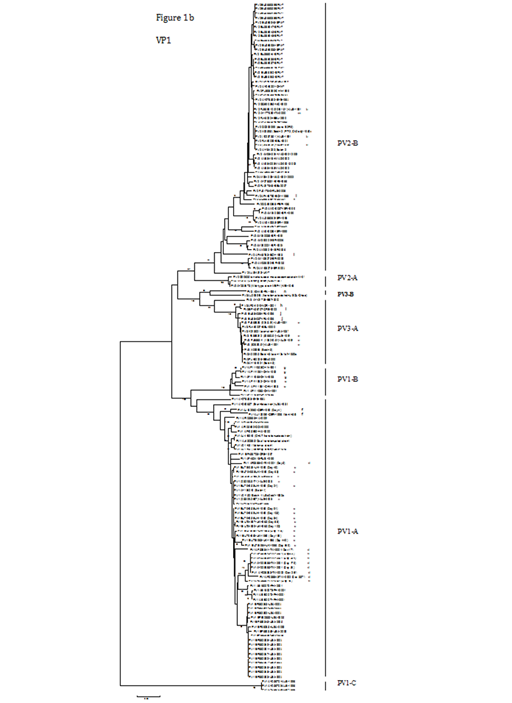
Figure 1B The corresponding VP1 groups or subgroups are indicated. Strain names indicate a GenBank accession no./country or area/year of isolation.
ALB, Albania; AUS, Australia; BEL, Belarus; BYE, Byelorussian Republic; CHN, The Chinese mainland; DOM, Dominican Republic; EGY, Egypt; EST, Estonia; FIN, Finland; GER, Germany; GRE, Greece; HAI, Haiti; IRA, Iran; ISR, Israel; ITA, Italy; MAD, Madagascar; ME, The Middle East; NIG, Nigeria; NOR, Norway; PER, Peru; PHI, Philippines; RUS, Russia; SPA, Spain; TW, Taiwan; UK, The United Kingdom
‘?’ indicates that the year of isolation was unknown.
The PV2 group in the VP1 tree (Figure 1b) could be divided into PV2-A and -B subgroups. PV2-A was composed of three strains, including a wild prototype PV2 strain (M12197), another wild-type strain (MEF-1) and an attenuated, mouse-adapted strain W-2, differing from each other by 0.4 to 1.6%. These three PV2-A strains showed 16.3 to 21.6% nucleotide difference in relation to the PV2-B strains. The large PV2-B subgroup encompassed the Sabin 2 vaccine strain (X00595 or AY184220) and other PV2 strains isolated from different parts of the world at different periods. Three local strains (labelled ‘b’) isolated in 1991 were clustered in this subgroup, with high nucleotide identity (99.2 to 100%) to the Sabin 2 strain (X00595). Several outbreaks of poliomyelitis associated with type 2 circulating VDPVs have been recorded; for instance, in Egypt from 1988 to 1993 (labelled ‘l’) (Yang et al., 2003) and in Madagascar (MAD) between 2001 and 2002.29 Moreover, from 1963 to 1966, widespread circulation of VDPVs took place in a region of Byelorussia (BYE).18 Another isolate (labelled ‘m’) was from an immuno deficient patient with poliomyelitis in Italy.30 Israeli (ISR) environmental surveillance of sewage from populations with high documented vaccine coverage (>95%) of confirmed efficacy identified two separate evolutionary clusters of type 2 VDPVs from 1998 to 2006.31
The PV3 group in the VP1 tree (Figure 1b) could be divided into two subgroups, PV3-A and -B. PV3-A consisted of the wild prototype PV3 strain (K01392), the Sabin 3 vaccine strain (X00925, AY184221 or X00596) and several PV3 strains from different countries, such as Norway (labelled ‘h’), Greece (labelled ‘i’), Estonia (EST)32 and Belarus (BEL). Four local strains (labelled ‘c’) from 1991 were also included in this subgroup, with high nucleotide identity (99.6 to 99.9%) to the Sabin 3 strain (X00925). Two strains (labelled ‘j’) were isolated from an Iranian child with X-linked a gammaglobulinemia.33 PV3-B included two strains; one (labelled ‘n’) was isolated during an outbreak of poliomyelitis in Finland in 1984,34 and the other one (AJ293918) caused the 1968 poliomyelitis epidemic in Poland.35 The relatively close relationship on the capsid coding region between these two strains was confirmed by a recent study.36
In comparison with the VP1 tree (Figure 1b) of PV1, -2 and -3, most PV strains showed concordant clustering in the 5’ UTR tree (Figure 1a), with some exchanges in tree branching for some strains. Variation in the 5’ UTR usually involved nucleotide substitutions or insertions/deletions. Three local PV1 strains (labelled ‘a’), three PV2 strains (labelled ‘b’) and four PV3 strains (labelled ‘c’) had high nucleotide identities in partial 5’ UTR sequences to their respective Sabin strains, ranging from 99.6 to 99.8%, 98.8 to 99.6%, and 99.6 to 99.8%, respectively. However, a few strains exhibited inconsistency in both trees. For example, two strains (AJ293918 and X04468-FIN-1984 [labelled ‘n’]) belonging to PV3-B in VP1 were separated in the 5’ UTR tree. In the 5’ UTR tree, the strains determined to be the most closely related to one of the PV3-B strains (AJ293918) were seven PV1-B isolates from the Chinese mainland, with nucleotide identity between 86.5 and 88.8%. Another five strains (DQ890386 [labelled ‘k’], AF448782 [labelled ‘l’], AF448783 [labelled ‘l’], AY560657 and AY278553), which located between PV1-B and PV2-A in the 5’ UTR tree, also displayed different clustering compared with their positions in the VP1 tree.
Data analysis for CVA11, -20, -21, -24 and PVs
The PVs are distinct from most of other members in EV-C only in the capsid region [29,37]. The phylogenetic relationships among 7 PV1, 6 PV2, 8 PV3, 4 CVA11, 14 CVA20, 15 CVA21 and 14 CVA24 strains including 16 local isolates, (Table 1) were inferred by the neighbor-joining method using the nucleotide/Kimura two-parameter method with MEGA, based on alignment of the partial VP1 and partial 5’ UTR nucleotide sequences (Figure 2).

Figure 2A Phylogenetic trees showing the genetic relationships among 7 PV1, 6 PV2, 8 PV3, 4 CVA11, 14 CVA20, 15 CVA21 and 14 CVA24 strains (including 3, 3, 4, 1, 2, 1 and 2 local isolates, respectively, [labelled ‘a’, ‘b’, ‘c’, ‘d’, ‘e’, ‘f’ and ‘g’, respectively], with laboratory identifiers provided in parentheses) based on alignment of the partial VP1 (322 to 328 nucleotides) and partial 5’ UTR (487 to 493 nucleotides) sequences. Trees were constructed by neighbour joining using Nucleotide/Kimura 2-parameter method with MEGA, version 5.0.21 Bootstrap values (percentage of 1,000 pseudo replicate data sets) of ≥75% supporting each cluster are shown at the nodes. The scale bars represent the genetic distance.
In the VP1 dendrogram (Figure 2b), all strains were monophyletic with respect to their homotypic prototype strains, and clusters were supported by high (>90%) bootstrap values. Strains generally segregated into seven distinct groups according to type (i.e., CVA11, -20, -21 and -24; PV1, -2 and -3). Several types, such as PV1, -2 and -3 (which were composed of respective wild prototype and Sabin vaccine strains, and 10 local PV strains [labelled ‘a’, ‘b’ or ‘c’]), formed relatively tight and well defined groups.
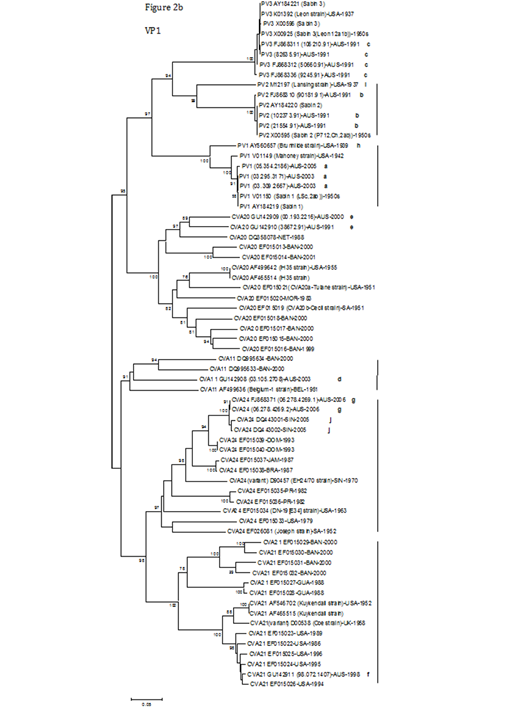
Figure 2B The corresponding VP1 groups are indicated. Strain names indicate a GenBank accession no./country or area/year of isolation.
AUS, Australia; BAN, Bangladesh; BEL, Belgium; BRA, Brazil; DOM, Dominican Republic; GUA, Guatemala; JAM, Jamaica; MOR, Morocco; NET, Netherlands; PR, Puerto Rico; SIN, Singapore; SA, South Africa; UK, the United Kingdom
Other types, such as CVA20 and -21, formed discernible subgroups, but still existed independently. CVA20 was divided into two subgroups; one was made up of five strains (two local strains [labelled ‘e’] and three from the Netherlands or Bangladesh), and the other subgroup consisted of nine strains (the prototype CVA20 IH35 strain, CVA20a-Tulane strain, CVA20b-Cecil strain and other strains from Morocco or Bangladesh). The partial VP1 nucleotide and amino acid identities of the two CVA20 local strains (00.193.2216/38672.91, labelled ‘e’) to the prototype CVA20 IH35 strain were 75.3/75.4% and 90.5/91.7%, respectively. CVA21 was also divided into two subgroups. The first consisted of six strains from Bangladesh and Guatemala; the second subgroup contained a local strain (labelled ‘f’), the prototype CVA21 Kuykendall strain, a variant strain (Coe strain) and another five strains isolated in the USA. The CVA21 local strain (labelled ‘f’) showed 90.3% partial VP1 nucleotide identity and 98.1% partial VP1 amino acid identity to the prototype CVA21 Kuykendall strain, and was closely related to five strains isolated in the USA from 1986 to 1996, differing from them by 1.4 to 3.2% of nucleotides.
CVA24 group contained two local strains (labelled ‘g’), the prototype CVA24 Joseph strain, the prototype CVA24 variant (EH24/70 strain), and other strains from Dominican Republic, Jamaica, Brazil, Singapore (labelled ‘j’), Puerto Rico and the USA. The partial VP1 nucleotide and amino acid identities of both CVA24 local strains (labelled ‘g’, with 100% VP1 nucleotide identity) to the prototype CVA24 Joseph strain were 77.4% and 91.5%, respectively. Furthermore, both CVA24 local strains had 97.7% and 98.0% VP1 nucleotide identities to another two CVA24 strains (DQ443001 and DQ443002, labelled ‘j’) isolated in Singapore in 2005,38 respectively. Compared to the other groups, the structure of CVA11 was uncomplicated, and only comprised four strains (a local strain [labelled‘d’], the prototype CVA11 Belgium-1 strain and two Bangladeshi strains). The partial VP1 nucleotide and amino acid identities of the CVA11 local strain (labelled‘d’) to the prototype CVA11 Belgium-1 strain were 78.9% and 92.7%, respectively.
As expected, in the 5’ UTR tree (Figure 2a), the vast majority of the strains belonging to their respective types were in different clusters except for the PV3 strains, which still formed a compact group. Ten local PV strains (labelled ‘a’, ‘b’ or ‘c’) were still positioned in the respective PV1, -2 or -3 group. Similar to the VP1 dendrogram, both CVA24 local strains (labelled ‘g’, with 100% partial 5’ UTR nucleotide identity) had 98.8% and 99.0% partial 5’ UTR nucleotide identities to two CVA24 strains from Singapore (DQ443001 and DQ443002, labelled ‘j’), respectively. As in the VP1 tree, the CVA21 local strain (labelled ‘f’) was also clustered tightly in the 5’ UTR with five strains isolated in the USA from 1986 to 1996, differing by 0.8 to 3.3%.
In addition, the prototype PV1 strain (AY560657, labelled ‘h’) was separated from the major PV1 group in the 5’ UTR tree, and was closely related to two CVA24 strains from the USA, differing by 12.1 to 12.4% of nucleotides. Similarly, the prototype PV2 strain (M12197, labelled ‘i’) was also segregated from the major PV2 group, and grouped with a CVA24 subgroup (containing two local strains [labelled ‘g’] and other strains from Dominican Republic and Singapore [labelled ‘j’]), with nucleotide identity to this subgroup ranging from 90.7 to 91.2%.
Evolutionary diversity of PV1, -2 and -3
Poliomyelitis was a seasonal disease, with peak transmission in the summer and autumn.39 Immunization with attenuated oral poliovirus vaccine (OPV) of Sabin is the cornerstone of the World Health Organization (WHO) program for the global eradication of poliomyelitis.
A common feature of the Sabin OPV strains is the presence of nucleotide substitutions in the IRES of 5’ UTR, which in serotypes 1 and 3 are critical attenuating mutations. Additional mutations encoding amino acid substitutions in the capsid region (including VP1) contribute to and stabilize the attenuated phenotype.39 For example, the single most important determinant of the attenuated phenotype of Sabin 1 was the A→G substitution at position 480 (abbreviated A480G) in the IRES.40 Four other substitutions (one in VP4, one in VP3 and two [G2795A and C2879U] in VP1) also contributed to the attenuated phenotype, and one substitution in the 3D polymerase region contributed to the temperature-sensitive (but not attenuated) phenotype.41-43 Three local PV1 strains (labelled ‘a’ in Figure 1) also showed nucleotide substitutions (A480G) in the 5’ UTR. In the VP1 region, two local PV1 strains (03.295.3171 and 03.309.2667) had G2795A substitutions, and one (05.354.2186) contained a ‘G’ at position 2795. In addition, all three isolates had C2879U substitutions in their VP1 regions.
For the Sabin 2 vaccine strain, one nucleotide substitution (G481A) in the IRES and another one (C2909U encoding a Thr→Ile substitution at position 143 of VP1) in VP1 appear to be responsible for the attenuated phenotype.44,45 Of three local PV2 strains, one (21554.91) showed a G481A substitution in the 5’ UTR; whilst another two (102373.91 and 90181.91) had a ‘G’ at position 481. In the VP1 region, two (102373.91 and 21554.91) showed substitutions (C2909U) and the third (90181.91) had a ‘C’ at position 2909.
Three substitutions (C472U in the IRES, C2034U in VP3 and U2493C encoding an Ile→ Thr substitution at position 6 of VP1) appear to be the main determinants of the attenuated phenotype of Sabin 3 vaccine strain.46-48 All four local PV3 strains (labelled ‘c’ in Figure 1) had a ‘C’ at position 472 in the 5’ UTR, consistent with the observed rapid reversion of this nucleotide in the human gut.46 In the VP1 region, one local strain (106210.91) had a substitution (U2493C) and three contained a ‘U’ at position 2493.
PV surveillance is important in the final stage of global polio eradication. From 2001, the WHO polio network laboratories routinely sequence the complete VP1 gene of all wild-type PV and VDPV isolates for comparison, based on the observation that VP1 genes encode several serotype-specific antigenic sites47 and evolve primarily by successive fixation of nucleotide substitution.39 Using VP1 sequence analysis, the genetic diversity of PV strains can be exploited in molecular surveillance.32 Moreover, the rapid evolution of PVs permits the use of comparative VP1 nucleotide sequencing to resolve the fine structure of a poliomyelitis outbreak, and to elucidate individual chains of transmission.50 As shown in Fig. 1, phylogenetic trees or lineage maps can be used for demonstration of sequence relationships.24,32,51
PVs are among the most rapidly evolving viruses known, with most evolution appearing to be random genetic drift, since >80% of nucleotide substitutions within the coding region generate synonymous codons. Most nucleotide substitutions of 10 local PV isolates (Figure 1b) existed at synonymous sites in the VP1 genes; however, some mutations were non-synonymous, which altered the amino acid sequences compared to the Sabin strains. For instance, at position 54 of the PV3 VP1 amino acid sequence, one isolate (9245.91, labelled ‘c’) had a Val instead of Ala in Sabin 3, whilst another two (106210.91 and 82635.91, labelled ‘c’) had a Thr (not shown). Nucleotide substitutions accumulate at an overall rate of approximately 1% per year at all sites, and at approximately 3% per year at synonymous sites,25,27,52 calculated primarily on the VP1 region or P1/capsid region. Evolution rates appear to be similar across serotypes and between wild-type PVs and VDPVs.24,52 The study on the evolution of Sabin 1 strains (labelled ‘e’ in Figure 1) in a hypo gamma globulinemic patient over 649 days demonstrated that, during the early period, the main driving force for genetic diversity appeared to be the selection of mutations at attenuation sites, particularly in the 5’ UTR and the VP1 BC loop.19
PVs recombine actively with each other and with other closely related EV-C members, a process which might offset the effects of accumulation of deleterious mutations arising in some lineages.25,36,53,54 In PV1-B of the VP1 dendrogram (Figure 1b), five (labelled ‘g’) of seven strains were type 1 (intra serotypic) wild-vaccine recombinant PVs, with a 367-nucleotide block of sequence (position 3271 to 3637) derived from the Sabin 1 strain spanning the 3’ terminal sequences of VP1 (115 nucleotides) and the 5’ half of 2A (252 nucleotides); however, another two strains (AF111983 and AF111984) were wild-type PVs. These recombinants are unusual because they involve a crossover within the VP1 capsid region. Homotypic natural infections involving different PV genotypes are presumably rare, except for those involving wild-type and VDPVs.27 Additionally, in PV1-A of the VP1 tree with the nine Taiwanese isolates (labelled ‘d’) collected over a 337-day period from an immuno deficient patient after onset of paralysis, relationships among the aligned complete sequences of the day 5, day 18 and day 52 isolates (i.e., different lineages) revealed a possible recombination site between nucleotide positions 2659 and 2678, near the 5’ terminus of the VP1 region.25
Natural interserotypic recombination within the capsid region appears to be rare, probably because structural incompatibilities restrict the fitness of most interserotypic recombinants.53 Some examples demonstrated the uncommon recombination within VP1. One strain (PV3 AF541919-NOR-2001) (labelled ‘h’ in PV3-A of Figure 1) was a Sabin3/Sabin 2/Sabin3 interserotypic recombinant PV, whose recombination junctions were at the 3’ end of VP1 region and in the 3D polymerase region.55 Another example is the strain (PV3 EF456707-GRE-2002) (labelled ‘i’ in PV3-A), whose Sabin 3/Sabin 2 recombination event was at the 3’ end of VP1 region.56 By contrast, interserotypic recombinants excreted by recent vaccines exposed to trivalent OPV and recombinants among circulating wild-type PVs and circulating VDPVs usually have crossovers restricted to the non capsid region.53,57 In this study, a few strains demonstrated significant incongruence of clustering in 5’ UTR and VP1 trees (Figure 1). For instance, recombination could explain the observation of one strain (PV2 DQ890386-NIG-2001) (labelled ‘k’) in the 5’ UTR tree that was separated from PV2-B in the VP1 tree: sequences upstream of nucleotide position 620 within the 5’ UTR and downstream of nucleotide position 5840 in the 3C region were derived from EV-C EVs.58 Another two strains (PV2 AF448782-EGY-1988 and PV2 AF448783-EGY-1993) (labelled ‘l’) in the 5’ UTR tree also suggested possible recombination. Complete genomic sequences of these two isolates revealed that their 5’ UTR and noncapsid-3’ UTR sequences were derived from other EV-C EVs.51 Moreover, the 5’ UTR and the non capsid region of the strain (PV3 X04468-FIN-1984, labelled ‘n’ in the 5’ UTR tree) appeared to be composed of variable-length stretches of nucleotides with closest resemblance to different EV-C strains.36 In fact, the recombination between PVs and other EV-C viruses is a normal feature of PV evolution in nature.
At present, three categories of PV isolates are recognized39
WHO declared the Western Pacific region, including Australia, polio free in 2000. Transition to exclusive use of inactivated polio vaccine (IPV) eliminates the risk of new cases of vaccine-associated paralytic poliomyelitis and new immuno deficient VDPV infections, and may maintain population immunity in many countries.39 As of November 2005, Australia moved to the exclusive use of IPV;60 therefore, PV strains are no longer isolated from clinical specimens in our laboratory. The local PV1 strain ([05.354.2186]-AUS-2005, labelled ‘a’ in Figure 1) was isolated just before OPV was stopped in Australia. However, the importation of wild-type PV1 from Pakistan to Melbourne, Australia, in July 2007 represented the first case of wild PV infection in Australia for 30 years,60 and highlights the need for continued vigilance for polio-like illness.
Evolutionary diversity of CVA11, -20, -21, -24 and PVs
CVAs of the EV-C species cause mild respiratory tract infections, and sometimes infantile diarrhoea, epidemic myalgia, infectious hepatitis, acute hemorrhagic conjunctivitis or poliomyelitis.1 The CVA24 variant, together with EV70, has caused several outbreaks of acute hemorrhagic conjunctivitis.38,61 CVA24 can occasionally be associated with acute flaccid paralysis,62 while CVA21 induces mainly mild respiratory infections and can be associated with encephalitis.63
EV-C isolates (excluding PVs) have been poorly studied due to the inability of certain serotypes (e.g., CVA1, -19 and -22) to grow in cell culture,37 the lack of specific antisera63 or the existence of antigenic variants. There are several examples of antigenic variants within an EV-C type, such as three CVA20 variants (CVA20a, CVA20b [Figure 2] and CVA20c) and the prototype CVA24 variant (EH24/70 [Figure 2]) isolated during an extensive epidemic of acute hemorrhagic conjunctivitis in 1970 in Singapore.64 However, analyses based on the comparison of VP1 nucleotide sequences with databases of EV prototype and variant VP1 sequences allowed us to search for such EV-C isolates. A partial or complete VP1 nucleotide sequence identity of ≥75% (>85% or 88% amino acid sequence identity) between a clinical EV isolate and a prototype serotype strain may be used to confirm the serotype of the clinical isolate, with the proviso that the second highest score is <70%.12,65 Three genotypes (CVA11, -20 and -21) were identified in our laboratory by partial VP1 sequencing of nonserotypeable isolates (Table 1).
The major worldwide CVA24 epidemics have been attributed to four genotypes of CVA24 variant strains (I to IV) on the basis of phylogenetic analysis of the 3C protease or VP1 region.66,67 In CVA24 variant strains, the estimated evolutionary rates are 1.0 - 4.1 × 10-3 or 6.27 - 6.67 × 10-4 per nucleotide per year based on the 3C protease gene,37,66,68,69 4.2 × 10-3 per nucleotide per year based on the VP4 gene,37 and 1.83 – 2.17 × 10-3 per nucleotide per year based on the 3’ VP1 gene.69
Interestingly, both local CVA24 strains isolated in 2006 (labelled ‘g’ in Figure 2) had similarly high nucleotide identities in both 5’ UTR (>98.8%) and VP1 region (>97.7%) to two other CVA24 strains (labelled ‘j’) isolated in Singapore in 2005.38 This temporal and geographical relatedness, combined with the phylogenetic analysis, suggests that the two local strains may have evolved from the CVA24 strains in Singapore before spread to Australia. Likewise, the local CVA21 strain isolated in 1998 (labelled ‘f’) was clustered closely in both the 5’ UTR and VP1 region with five strains isolated in the USA from 1986 to 1996, suggesting the possibility of a North American origin.
The PVs are distinct from most of other members in EV-C only in the capsid region, suggesting that recombination has played a major role in their evolution.29,37,54 Recombination may lead to a lack of linkage between non capsid sequences and the neutralization serotype of a virus within a species.37 In EV-C, a virus with a PV capsid may recombine with a CVA to acquire different non capsid sequences; in turn, a virus with a CVA capsid may acquire different non capsid sequences by recombination with a PV. For example, the most striking feature of the CVA21 variant strain (Coe strain, Figure 2) is the remarkable homology to the PVs (>90% at the amino acid level) in the 3’ part of the genome, suggesting that it is a recombinant virus.70 The 5’ UTR phylogenetic tree demonstrated disordered topology compared with the VP1 dendrogram (Fig. 2), indicating that the 5’ UTR of these types has frequently been subject to recombination.29
Evolutionary diversity of EV-C
Molecular typing approaches (including PCR and sequencing) allow the rapid and accurate identification of EV-C viruses. Phylogenetic analysis of sequences is the best method to discriminate between variants within a type, to confirm the common source of isolates during an outbreak, and to study the evolution of a given type or among different types.14 The genetic comparison of different EV genes (such as 5’ UTR and VP1) provides strong evidence of epidemiological linkage of EV-C strains in some types (such as PV1, -2 and -3 in Fig. 1; CVA21 and CVA24 in Figure 2). The combination of molecular typing and phylogenetic sequence analysis will guide individual patient diagnosis and benefit the investigation of EV evolution.
The intratypic genetic variation of the 5’ UTR and VP1 regions in EV-C viruses reveals that distinct genome regions evolve differently, reflecting their different roles. Structural integrity of the 5’ UTR is fundamentally important for efficient viral replication and for virulence,71 while the VP1 capsid protein contains most neutralization epitopes, and VP1 sequence correlates well with antigenic typing by neutralization. In the VP1 region, EV-C types cluster into three subgroups9,37 which are designated as A to C. CVA21 and -24 belong to subgroup B, while PV1 to -3 and CVA20 are included in subgroup C. CVA-11 groups together with the strains of subgroups B or C depending on the method of phylogenetic tree construction.9
Mutation and recombination are important in EV evolution and diversity. Studies of EVs suggested that sequence variations in the UTRs, the VP1 region and the 3D polymerase could affect virus infection and replication capability in vitro and in vivo.72,73 Base miss incorporation during chain elongation and lack of 3’ to 5’ exo nuclease proofreading ability in RNA polymerases can cause very high error rates; there is a spontaneous mutation rate of approximately one mutation per genome per replication.74 The evolution of EVs occurs through genetic drift and, over much longer periods, antigenic diversification in the structural gene region encoding the virus capsid (including VP1).75 Studies have shown that EV 5’ UTR mutations markedly decrease multiplication efficiency,76 alter cell tropism,77 and attenuate virulence.78,79 The mean nucleotide substitution rates of EV-C subgroup B (ranging from 1.170 to 3.625 × 10-3 with combined high-probability distribution interval of 1.170 to 3.625 for partial VP1) are slightly lower than the corresponding estimates for other EV types. Intratypic genetic change of VP1 is dominated by silent mutations accompanied by amino acid polymorphism occurring dominantly at immunogenic sites, while intertypic differences of VP1 include permanent fixation and insertion/deletion of distinct ‘signature’-amino acids that could be a result of larger scale changes in the capsid structure.9 In this study, the relatively high sequence diversity among the 5’ UTR and VP1 regions of EV-C viruses suggests that nucleotide substitution is probably the dominant evolutionary mechanism in these regions of the genome.
Genetic recombination of EVs was first described with PVs.80 The majority of recombination occurs within P2 and P3 regions, with little or no recombination within the P1.81 Generally, only the capsid P1 region (especially VP2-VP3-VP1) is inherited as a single unit, suggesting that the capsid is the primary determinant of EV identity.10,82 The frequency of recombination differs between EV species, being higher in EV-B species than in EV-A species83 and very low in EV-D species.84,85 The evolutionary diversity of EV-C is also due to intratypic/ intertypic (or interspecies) recombination between the structural and non-structural coding regions and the 5’ UTR.29,37,86,87 In particular, the prototype EV-C strains are recombinant relative to each other,37 and circulating EVs commonly recombine with wild-type and vaccine PVs.53 The phylogenetic pattern of the EV-C strains in a recent study suggests non-random recombination between strains of the same species. The frequency of recombination appears to differ between species, types and intra species groupings, which is possibly due to cell tropism, sequence similarity and the phylogenetic position of a given virus strain.86 In this study, 5’ UTR phylogenetic trees from EV-C showed apparent non-convergence among respective different sequences within many types, compared with VP1 trees (Figure 2), indicating that intertypic recombination may play a major role in the evolution of some of these viruses. Moreover, PVs appear to recombine actively with each other and with other closely related EV-C members (Figures 1 & 2).
This study focuses on molecular sequence analyses of the 5’ UTR and VP1 regions of 199 EV-C strains. Understanding the tempo and pattern of molecular diversity and epidemiology is of great importance in the pathogenesis of EV-C Enteroviruses, information which will assist in disease prevention and control (including PV infections).
In conclusion, EV-C viruses evolve quickly, mainly due to high mutation and recombination rates. The 5’ UTR is fundamentally important for efficient viral replication and for virulence; the VP1 region can be used for virus identification and evolutionary studies. The sequence analyses of the 5’ UTR and VP1 regions of 199 EV-C strains demonstrated that
We thank Kenneth McPhie for assistance with provision of clinical samples.
None.

©2015 Zhou, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.