Journal of
eISSN: 2373-6453


Research Article Volume 3 Issue 3
1National Centre of Excellence in Molecular Biology, University of the Punjab, Pakistan
2Department of Microbiology, Baluchistan University of Information Technology, Engineering and Management Sciences, Quetta
Correspondence: Abrar Hussain, Department of Microbiology, Balochistan University of Information Technology, Engineering and Management Sciences, Airport Road, Baleli, Quetta, Pakistan, Tel 92 (81) 2880410, 92 (81) 2881036
Received: April 03, 2016 | Published: April 19, 2016
Citation: Khubaib B, Idrees M, Hussain A (2016) Construction of Mammalian Expression Vector of Core Gene of HCV of Pakistani Isolate Genotype 1a. JHum Virol Retrovirol 3(3): 00089. DOI: 10.15406/jhvrv.2016.03.00089
Background: Cell lines are a valuable tool to identify the HCV virus infection and propagation and by establishing cell lines expressing core protein we can explore the role of HCV core protein in development of HCC.
Results: The results indicate PCR amplification of 573bp product of core gene and sequencing confirmed the 1A genotype of HCV of Pakistani isolate. Then recombinant plasmid was digested with restriction enzymes (Hindi III and EcoR I) which gave fragments of 5.5 kb and 0.6 kb and It was a prove that our plasmid having a core gene. 21KDa core protein extracted from the transiently transfected Huh 7 cell line with recombinant mammalian vector of core gene was detected by using anti-core monoclonal antibody.
Conclusion: we successfully constructed a mammalian expression vector of core gene of Pakistani isolate of genotype 1A that was encoding 21KDa of core protein in Huh 7 cell line.
Keywords:Chronic hepatitis, HCV, vaccine, HCC, DNA, Hepatocellular carcinoma HCC
HCV, Hepatitis C Virus; ABI, Applied Biosystem Inc; RT-PCR, Reverse Transcriptase Polymerase Chain Reaction; cDNA, Complimentary DNA, MMLV, Moloney Murine Leukemia Virus; HCC; Hepatocellular Carcinoma; SST, Serum Separation Tubes
Chronic hepatitis is mostly caused by Hepatitis C virus and it is reported that 170 million people worldwide are infected with this virus1 that frequently leads to hepatocellular carcinoma in 10% to 80% patients in different populations. In Pakistan, HCC is a leading cause of death and accounts for 60%-90% of all primary liver malignancies.2
In 1989, HCV was identified, a positive sense, single stranded RNA enveloped virus in length of 9600 nucleotides that encodes a polyprotein with 3 structural (Core, E1, E2) and 7 nonstructural (p7, NS2, NS3, NS4A-B, and NS5A-B) proteins.3-4
Molecular events that develop HCC during HCV infection are feebly identified. In this regard, HCV core protein takes an important part in the development of Hepatocellular carcinoma and steatosis in transgenic mice.5-6 Core gene is highly conserved region in different genotypes of HCV so it is an ideal candidate for DNA vaccine development.7 Protein encoded by it is extremely basic in nature that forms the nucleocapsid of virus. At its N terminal Core protein of HCV is rich in arginine and lysine amino acids and it binds with RNA in order to form an envelope of HCV.8
Although main function of core protein is in genome packaging, but its sub cellular localization reveals that it modulates numerous cellular processes.9 Localization of core protein into mitochondria indicates that it modulate apoptosis and lipid transfer.10 In cytoplasm, it accumulated on lipid droplets and co localized on apolipoprotein AII shows its dealing with cellular lipid metabolism.11 When hydrophobic part at C-terminal of core protein is deleted, it translocated in nucleus12 and interacted with several cellular proteins, like cytopalsmic tail of lymphotoxin-β receptor,13 retinoid X receptor-α, type 1 TNF-α receptor and nuclear ribonucleo-protein K.14
HCV core protein is multifunctional protein it modulate multiple intracellular signaling pathways like proliferation of cell, apoptosis, cell cycle, various oncogenes and growth factors and stimulate humoral and cellular immune system.15 It is involved in plasmacytoid dendritic cells apoptosis and inhibition of IFN-α production,16 suppress the p53 transcription,17 repress the phosphorylation and DNA binding property of stat 3,18 and suppresses cell proliferation and apoptosis by preventing the phosphorylation of MAPKs and stimuating the transcriptors NF-kappaB and AP-1, thus endorse the HCV persistent infection leads to chronic hepatitis C disease and hepatocellular carcinoma.19
The aim of the study was to construct a recombinant mammalian expressing vector of core gene of Pakistani isolate of HCV genotype 1A and to establish an experimental model for exploring the role of HCV core protein in development of HCC.
Sample collection and RNA isolation
Patients era infected with Hepatitis C virus, genotype 1A, were obtained from Molecular Diagnostics lab, CEMB, University of the Punjab Lahore by using BD Vacutainer collection tubes (Becton Dickenson). For isolation of serum, serum separation tubes (SST) were used. The serum was recovered after centrifugation at 2000g for 10 minutes. RNA was isolated from 100µl of serum, using commercially available Gentra RNA isolation kit (Puregene, Minneapolis, MN 55441 USA), according to the kit protocol with little modification.
cDNA synthesis and PCR amplification:
Reverse transcription (RT) was performed by incubating the 10μL of RNA used as template in a 10-μL reaction mixture having 10 picomole (pmol) of core specific antisense primer COAS (Table 1), 100 units of MMLV reverse transcriptase (invirtogen), 10 units Ribolock RNase Inhibitor (Fermentas), 1X First Strand Buffer (invitrogen), and 10 mM deoxynucleotide triphosphates (dNTPs) at 37oC for 50 minutes, 42oC for 10 minutes, 95oC for 3 minutes and 20oC for 2 minutes. PCR Amplification of core region of HCV was done by using 4µl of synthesized cDNA as template with outer forward COS and reverse COAS (Table 1) primers. Then 2 µl of first round product was taken as template and re- amplified by performing nested PCR with internal forward CIS (Table 1) and reverse CIAS primers. The amplified PCR product was run on 2% agarose gel having ethidium bromide and visualized under a Ultra Violet transilluminator. The amplified product of 573bp was purified by using Bead DNA Gel Extraction Kit (Fermentas CAT# K0513).
Sequencing PCR
Amplification of core gene of genotype 1A was confirmed by sequencing that performed by using ABI PRISM 3100 Genetic Analyzer (Applied Biosystem Inc., Foster City, CA, USA) in both directions.
TA cloning
Purified core gene ligated into TA vector by using TA cloning kit (Cat# K2020-20, Invitrogen) and the manufacturer’s protocol was followed. The ligated product was transformed into E.coli strain TOP10F and competent cells that have taken up recombinant plasmid are then selected onto a Luria-Bertani agar plate having 100 μg/ml of ampicillin and 12.5 μg/ml of tetracyclin. Colony PCR was performed with two sets of primers (gene specific primers CIS, CIAS and T/A vector specific primers). Then recombinant plasmid, named as PAK-C1A, was digested with restriction enzyme ECOR I and digested DNA was checked on 1% agarose stained with ethidium bromide under U.V illuminator.
Construction of expression vector
The PAK-C1A plasmid containing core region was used as template for PCR amplification of core region. A set of restriction primers RCIS and RCIAS (Table 1), having restriction sites for enzymes HindIII and EcoR 1 in the inner set of sense and antisense primers at 5` site respectively, was used for PCR amplification. The amplified product was digested with enzymes and recombined into pcDNA3.1 (+) mammalian expression vector (Invitrogen). The recombinant was transformed into E.coli strain TOP10F and positive colonies were cultured in Luria-Bertani medium (LB media) having 100 μg/ml ampicillin and 12.5 μg/ml of tetracyclin. In order to confirm the fragment insertion into vector Colony PCR was performed with two sets of primers, gene specific (CIS & CIAS) and vector specific (T7 & BGH) primers. 1.5% Agarose gel electrophoresis was run to visualize PCR product. The recombinant mammalian expression vector, named as pcDNA-PKcore, was extracted using GeneJet Plasmid Miniprep Kit (Fermantas CAT # K0503) and further confirmation was done by restriction analysis. The proper orientation core gene of HCV encoding protein was confirmed by sequence analysis.
Huh 7 cell line and transient transfection
Huh-7 human liver hepatoma cell lines (American type cell culture) were grown in Dulbeccos modified Eagles medium (ICN technologies USA) have 100 ug/ml streptomycin and 100 U/ml penicillin and 10% Fetal bovine serum (FBS) (Gibco life science technologies USA). Three days before transfection cells were seeded onto 60mm petri plate and when cells became 70-80% confluent then transient transfection was performed using Lipofectamine-plus 2000 reagent (Invitrogen), following the manufacturer`s protocol.
Reverse transcription (RT) PCR analysis
To characterize the core producing cell, we have screened the expression profiles of cell line by RT-PCR. After 24 hours of Transfection, total cellular RNA was extracted using RNA Isolation Kit (Gentra biotronics USA) and rehydrated into 20 µl of RNA hydration solution for 30 minutes at 4ºC. Then 10µl of this hydrated RNA was used for RT-PCR using antisense primer CIAS. The product of RT-PCR was subjected to PCR amplification using primers CIS and CIAS.
Western blot analysis
Protein was extracted from Huh 7 cells transiently transfected with recombinant and control plasmid. Media was removed from the transfected plates and cells were trypsinized for 2-3 mins to detach the cells from the surface. Then 1000 μl of DMEM media was added in the plate and mixed well to resuspend the cells in the media. Cells were shifted into a new sterile eppendorf and spinned for 3 minutes at 8000 rpm. Cells were washed with 500 μl of 1X PBS, resuspending the cells and centrifuged for 3 minutes at 8000 rpm. Supernatant was discarded and 60 μl of lysis solution, containing 150mM NaCl, 5mM EDTA, 1% triton 100, 0.01M tris HCl (pH 7.4), 0.1mM PMSF and Proteinase inhibitor cocktail, was added to the pellet and kept on ice for 10 minutes. Then centrifuged (13000 rpm) at 4°C for 10 min and supernatant was shifted to a new sterile eppendorf. Mixed with 45 μl of 2X sample loading dye and heat shocked in boiling water for 10 minutes and immediately transferred on ice for 5 to 10 minutes. Then SDS-PAGE was performed and protein was transfer onto nitrocellulose membrane. The membrane was incubated into 5% skim milk (blocking solution), then overnight incubated at 4ºC into primary monoclonal anti mouse antibody (Santa Cruz Biotechnology). Then washed with 1x PBST and incubated in secondary antibody, conjugated with alkaline phosphatase (AP), for 1 hour at 37ºC. Then HCV core protein was detected by chemiluminescence using NBT-BICP tablets (Sigma).
PCR amplification
The cDNA was synthesized using gene specific outer anti sense primer COAS (25pm). Then nested PCR was performed using two sets of primer outer set COAS and COS and inner set CIAS and CIS of primers (Figure 1). The PCR products obtained after amplification were run on the 1.5% agarose gel and stained with ethidium bromide (0.5 mg/ml) and were visualized by UV transillumination. The 573-bp fragments produced by PCR were identified by comparison with a molecular mass marker (Figure 2).
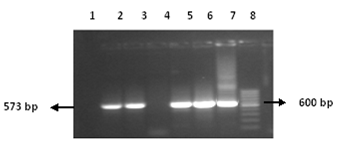
Figure 2 PCR Amplification of HCV core gene. Lane 2-3 & 5-7: bands of core gene, Lane: 1&4 negative control. Lane 8: 50 bp marker.
Sequencing PCR
Identification of amplified product was confirmed by sequencing PCR. Sequences of the core gene of HCV genotype 1A of Pakistani Isolate were searched for homology with other sequences in GeneBank and submitted to NCBI Genbank data base. The assigned Accession numbers for the local core gene sequence is GQ451336.
TA Cloning Of HCV Core Gene PCR Results:
The freshly eluted PCR product was ligated in the TA vector. The ligation reaction was used to transform into E.Coli strain TOP10F. Competent cells that have taken up plasmid were selected by spreading the culture onto a Luria-Bertani (LB) agar plate containing 100μg/ml of ampicillin and 12.4 μg/ml of tetracyclin. Colonies were selected by incubating the plate overnight at 37°C. To identify bacteria harboring cloned core gene, individual colonies were used to directly inoculate for colony PCR reactions using gene specific primers (Figure 3a). The results of vector specific primers counter proofs the presence of genes of interest in the TA vector as these primers originates from the outer regions of the cloned gene and PCR product of vector specific primer approximately 769 bp (Figure 3b). The resulting colonies were further analyzed by restriction digestion (Figure 3c) and DNA sequencing which shows homology with previously submitted sequence.
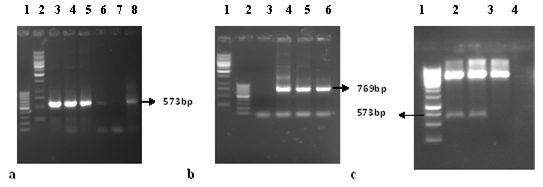
Figure 3 A. Colony PCR for screening of positive clones: Lane 1, 100bp marker. Lane 2, 1kb marker. Lane 3-8, Core Encoding clones with gene specific primers. b. Colony PCR for screening of positive clones: Lane 1, 1kp marker. Lane 2, 100bp marker. Lane 3-6, Core Encoding clones with vector specific primers. c. Gel picture for restriction digestion of core gene coding TA vector: Lane 1, 1kp marker. Lane 2 and 3, show digested TA vector with required size of gene. Lane 4, shows digested TA vector but do not have gene.
Cloning of HCV Core Gene in mammalian expression Vector Pcdna3.1
In order to characterize the role of core gene in disease progression we cloned the amplified PCR product in mammalian expression vector pc DNA 3.1(+). The vector has a CMV promoter which represents an effective mean to transduce eukaryotic cells for transient and stable expression studies (Table 1). The amplified sequence encoding core gene was digested, purified and cloned in expression vector between Hindi III and EcoR I sites. In order to clone CORE gene into pcDNA it is nessecary to amplified the core gene by using restriction primers RCIAS and RCIS and then ligated into pcDNA. Positive clones were used for amplification of the cloned core genes by using gene specific (CIS & CIAS) and vector specific primers (T7 and BGH) (Figure 4A). The results of vector specific primers counter proofs the presence of genes of interest in the expression vector as these primers originates from the outer regions of the cloned genes. Then recombinant plasmid was digested with restriction enzymes (Hindi III and EcoR I) which gave fragments of 5.5 kb and 0.6 kb (Figure 4B). It was a prove that our plasmid having a core gene. The presence of ATG, a start codon required for initiation of translation of protein, was confirmed by sequence of the pcDNA-PKCore plasmid.
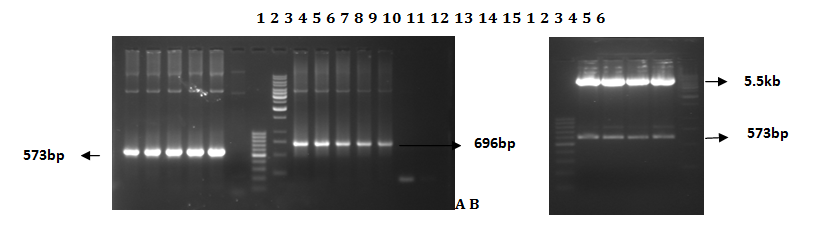
Figure 4A A.Colony PCR for screening of positive clones: Lane 1-6, Core Encoding clones with gene specific primers, Lane 7, 100bp ladder. Lane 8, 1kb ladder. Lane 9-14, Core Encoding clones with vector specific primers. Lane 15, –ve Control. B. Gel picture for restriction digestion of core gene coding pcDNA-PKCore vector: Lane 1, 100bp marker. Lane 2 to 5, show digested vector with required size of gene. Lane 6, 1kp marker.
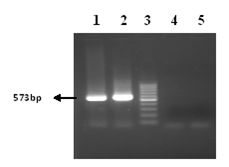
Figure 4B RT-PCR was done to characterize the core producing cell. Lane 1-2, RT-PCR transfected cells, lane 3 100bp marker & lane 4, RT-PCR of transfected cells with control plasmid. Lane 5, RT-PCR of untransfected Huh 7 cell line as a negative control.
Expression of core protein in transfected Huh 7 CELLS
Huh 7 cells were transiently transfected with core expressing vector pcDNA-PKCore and protein extraction was done from cells after 48 and 72 hours. Control plasmid pcDNA 3.1 (+) was also used to transfect the cells. Expression of HCV core proteins coded by expression vector was checked with protein extracts of transfected Huh-7 cells using mouse monoclonal sera against core (Santa Cruz Biotechnologies Inc. USA). The Western blot analysis identified specific bands of the expected electrophoretic mobility for core having molecular weight of 21 kDa (Figure 5). These results shows that Huh 7 cells transfected with the pcDNA-PKCore could express HCV core protein.
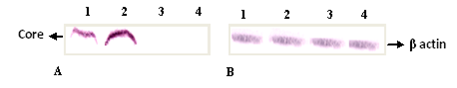
Figure 5 A. Western blot of core protein of HCV, detected with the anti-core antibodies. Lane 1: core protein at 48 hr post transfection, lane 2: core protein at 72 hr post transfection and Lane 3: protein extracted from transfected cells with control plasmid, lane 4: negative control, protein extracted from untransfected huh 7 cells. B. western blot of β actin, protein expression of a house keeping gene, was used as a control. Lane 1: at 48 hr post transfection, lane 2: at 72 hr post transfection and Lane 3: protein extracted from transfected cells with control plasmid, lane 4: negative control, protein extracted from untransfected huh 7 cells.
HCV core protein acts as multifunctional protein and at molecular level , it constitutes the virus capsid,20,21 associates in HCV pathogenesis,22 involves in hepatocellular fibrosis, steatosis and carcinogenesis,23 shows significance in HCV diagnosis24 and as most conserve region of HCV in all genotypes it is an important candidate gene for DNA based vaccine development against HCV infection25 all these functions indicates its critical role to induce HCV viral hepatitis that develop into carcinoma.
Although hepatitis C virus cause the persistent infection but to sustain the viral replication in vitro system of cell culture is very difficult and lack of small animal model is the major hinder to explore the HCV replication and propagation. Other than human the only hosts of HCV, are chimpanzees26 and small animal model, trimera mice.27 In this regard, cell lines are a valuable tool to identify the virus infection and propagation.28-29
In this study we constructed a recombinant mammalian expression vector of HCV core gene by using molecular cloning techniques that efficiently expressing core protein in Huh cell line. HCV RNA of Pakistani isolate genotype 1A was extracted from serum of HCV infected chronic patient and core specific primers was used for nested PCR amplification of core. Then amplified product was ligated into T/A cloning vector, PAK-C1A, which further used for amplification of core gene with restriction primers. Amplified product after restriction digestion was ligated into pcDNA 3.1(+) and recombinant plasmid pcDNA-PKCore was obtained, restriction analysis and sequencing PCR was used to identify newly constructed plasmid. Then it was used to transfect the 70-80% confluent Huh 7 cells with lipofectamine reagent. RNA expression of core gene in transfected cells was confirmed by performing RT-PCR after 24 hours of transfection and western blotting confirmed the successful expression of core protein after 48 hours and 72 hours transiently transfected cells. Our results show the successful cloning and expression of mammalian expression vector of core gene of HCV.
We conclude that newly constructed mammalian expression vector, pcDNA-PKCore, is efficiently transcribing RNA and encoding core protein in Huh cell line. That, pcDNA-PKCore, can be used for establishing the stable cell lines expressing HCV core protein which may be helpful to explore the molecular events that are affected by core protein in cells.
None.
None.

©2016 Khubaib, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.