Journal of
eISSN: 2373-6453


Research Article Volume 8 Issue 3
Centre for Interdisciplinary Research in Basic Science, Jamia Millia Islamia, India
Correspondence: Dr Fareeda Athar, Associate Professor, Centre for Interdisciplinary Research in Basic Sciences, Jamia Millia Islamia, New Delhi-110025, India, Tel +91-11-26981717 Ext. 4492, Fax +91-11.26980164
Received: May 20, 2020 | Published: July 22, 2020
Citation: Beg MA, Athar F. Computational method in COVID-19: Revelation of preliminary mutations of RdRp of SARS CoV-2 that build new horizons for therapeutic development. J Hum Virol Retrovirolog. 2020;8(3):62-72. DOI: 10.15406/jhvrv.2020.08.00223
COVID-19 struck the population with fear of infection with this non treatable disease. This is a consecutive infection of SARS superfamily infection after SARS and MERS infection that was occurred in 2003 and 2014 respectively. WHO named this virus as SARS CoV-2 and the disease caused by this infectious virus was introduced as COVID-19. The virus infects 1,000,889 humans all over the world out of which 210,244 recoveries and 51,371 deaths had been reported till 02nd April 2020. Herein, we are using various computational methods such as EASE-MM, PROVEAN, iSTABLE, STRUM, DUET, SDM, DynaMut and MAESTROweb to identify the effects of protein structure stability of NSP12 protein. We have identified 12-point mutation where the deleterious effect is in "high-confidence". Further analysis of these high-confidence point mutations demonstrates that the mutation in W107, W159 and F636 with Glycine has a highly deleterious effect on the structural stability of NSP12. This analysis provides a detailed understanding of the structural changes of NSP12 and the effect of different point mutations on structural stability variations. As NSP12 gene is an important part of RNA dependent RNA polymerase (RdRp) which is a part of active machinery for translation, a point mutation in the various residue of this gene might provide some crucial information about its functioning. We are dealing with the situation where we urgently required medication or vaccine to combat COVID-19. Therefore, this study for the first time provides essential molecular information about NSP12 gene and its significance in designing new therapeutic against COVID-19.
Keywords: COVID-19, NSP12 (RdRp), point mutation, structural stability, insilico approaches
MERS, Middle East Respiratory Syndrome; NCBI, National Centre for Biotechnology Information; SARS CoV-2, severe acute respiratory syndrome Coronavirus 2; RdRp, RNA dependent RNA polymerase; CoVs, Coronaviruses; NSP, non-structural protein; NSPs, non-structural protein sequences; RTC, replicase-transcriptase complex; +ssRNA, single stranded RNA of positive sense; kD, Kilodalton; PDB, protein data bank; 3D, three dimensional
A novel Coronavirus creates a major pandemic nowadays that cover almost the whole world in a lethal situation. This is the third serious pandemic due to coronavirus after severe respiratory syndrome (SARS) in 2003 and Middle East Respiratory Syndrome (MERS) in 2014.1 The novel coronavirus has named as SARS CoV-2 by WHO and the disease was described as COVID-19.2 The outbreak had started in a seafood market located in Wuhan, China and thought to be mainly spread through bats. As it is the recent epidemic, there is not much information is present for this virus. A Coronavirus is a large group of coronaviridae family that infects birds, animals and humans. CoV 229E, NL63, OC43, and HKU1 are some known non-severe acute respiratory syndrome (SARS)-like CoVs and not cause ant severe disease and described as endemic viruses. But the past two decades history comprised of severe and lethal disease caused by SARS and MERS coronavirus.3–5 The virus infects 1,000,889 humans all over the world out of which 210,244 recoveries and 51,371 deaths had been reported till 02nd April 2020.6 These viruses are positive sense enveloped viruses of largest genome size 30 kb among all single-stranded RNA of positive sense (+ssRNA). The genome comprises of 5’ cap and 3’ poly A tail that makes it perform as mRNA for translation of the replicase polyproteins. The replicase proteins mainly encode non-structural proteins (NSP) which covers almost 2/3rd of the total viral genome (20kb) and the rest is covered by structural proteins (10kb).7 The organization of the coronavirus genome is 5′-leader-UTR-replicase, S (Spike), E (Envelope), M (Membrane), N (Nucleocapsid), 3′UTR poly (A) tail with accessory genes scattered within the structural genes at the 3′ end of the genome. The S protein is the main protein that is utilized for viral entry into the host cell. S protein is almost 150 kD and comprises of an N terminal signal sequence to gain access to the ER in N glycosylated manner.8,9 This protein is present on the viral surface in a trimeric form that upon entry in the host cell surface, get cleaved by host cellular proteases S1 an S2. S1 participates in the attachment whereas S2 mainly provides structural support to the virion. The non-structural part (58%) of the virion is more conserved than the structural part (43%) among different coronavirus species. This is one reason for the frequent occurrence of mutation in these viruses but a second and major reason is its huge genome size which is usually less than 10kb in comparison to other RNA and DNA viruses.10
The replicase gene encodes two large ORFS, Replicase (rep1a) and Replicase (rep1b), which express two co-terminal polyproteins, pp1a and pp1ab. Polyproteins pp1a and pp1ab contain the NSPS 1–11 and 1–16, respectively.11,12 In pp1ab, NSP11 from pp1a becomes NSP12 following extension of pp1a into pp1b. Many of these NSPs work in a coordinate manner to systemize replicase-transcriptase complex (RTC) to generate an environment suitable for RNA synthesis, and eventually are responsible for RNA replication and transcription of the sub-genomic RNAs. Among all NSPs, NSP12 is an important gene that codes RNA dependent RNA polymerase (RdRp) gene.13,14
We are trying to elaborate on important aspects about NSP12 gene in this manuscript and the impact of different point mutations based on sequence and structure. These mutation studies give us essential information about the functioning of NSP12 and so replication machinery. Different point mutations also evaluate the functioning and essentiality of a specific portion of NSP12. The manuscript clarifies the impact of mutations on the structure which is an essential step to understand its pathogenesis and thus also helpful in understanding the disease.
Complete genome sequence of SARS CoV-2 is available in the National Centre for Biotechnology Information (NCBI) with the accession no. (NC_045512.2) was used for homology modelling.15 The homology modelling is done by using the SWISS-MODEL server which is freely available, and it builds the protein model with the given targeted-template sequence alignment. The NSP12 of SARS-CoV is 97% identical with the SARS CoV-2 (orf1ab gene sequence). The identical sequence template PDB ID: 6NUR was found which is 97.05% like NSP12.16,17 The predicted 3D model structure validation was done by SAVES metaserver of the University of California Los Angles (UCLA) where some server RAMPAGE, Verify3D and ERRAT which predict different stereochemical properties.18–21
The mutational consequence on protein by single point mutation by the Insilico methods was done by EASE-MM server, PROVEAN server, iSTABLE metaserver, STRUM server, DUET server, SDM server, DynaMut server and MAESTROweb which are online servers and done mutation based on protein sequence or protein structure.22
How to select the residues for point mutations?
The first step, for doing the single point mutation for selecting the single amino acid residue which is done by EASE-MM server. This server finds out the protein stability by changing the single point mutation for using the protein sequence. EASE-MM stands for Evolutionary Amino acid and Structural Encoding with Multiple Models which is based on support vector machine (SVM) models generated a Pearson correlation coefficient of 0.53–0.59 in the validation of 10-fold increase and free testing. At the point when distinguished with structure-based energy capacities, EASE-MM accomplished an equivalent or better execution.23,24
Sequence-based analysis of point mutations
PROVEAN is the software tool which uses the alignment-based score approach to generate the prediction for a single or multiple amino acid substitutions, in a frame of insertion or deletions. PROVEAN stands for Protein Variation Effect Analyzer and based on BLAST hits by CD hits a limitation of 75% worldwide succession identity and top 30 selected for further prediction and the score is expected to have a “deleterious” impact or a “neutral” impact.25,26 After the outcome of the point mutations, analyses those selected amino acids for iStable server which is the sequence-based mutational analysis tool.27 iStable server is inbuilt with an internal thermodynamic parameter which works by the variation of single point mutation. This server based on SVM method and work like a web computing manner which constructed via sequence info and estimate the results form divergent element forecaster.28,29 This server has 2 more outcome results shown iMutant2.0 and MUpred which compare the iStable conclusions. The outcome of the iMutant2.0 is in ΔΔG which measure the Gibbs free energy (ΔΔG) between wild-type and mutant. A mutant protein is recognized to destabilize if the ΔΔG value esteem <0 and stabilizing if the ΔΔG esteem>0. The MUpred predicted the confidence score is specified in the scope of -1 to 1 and the score is lesser than zero it means diminishing the stability and more than zero is vice versa.30,31 Based on sequence and structure-based method discriminated the analysis of point mutations was done by the STRUM server. STRUM server predicted the change of the protein stability by the sequence and structure based upon ΔΔG upon single point mutation and based on Pearson correlation coefficient (PCC) between predicted and measured changes of Gibbs free-energy gap. STRUM was done sequence profile score with the distinctive algorithm for numerous sequence alignment. The ΔΔG value is not vulnerable to the eminence of a structure made as much as the global fold is correct (TM-score > 0.5) and the ΔΔG> 0 shows stable conformation whereas ΔΔG< 0 is vice-versa.32
Structure-based analysis of point mutations
Protein structure-based mutation provides energy change value that is more defined towards its functionality. Mutations at sequence and structure level also help in differencing the impact of mutations at two stages as the primary sequence of protein must be travelled from various parameters of folding and unfolding to complete its final structure. This article emphasizes three in-silico tools (SDM, DUET, DynaMut) that had been utilized for mutation investigation depend on the structure.33 Site-Directed Mutation (SDM) Focuses on the effects of mutation on the structure of protein and function. Site-Directed Mutation (SDM) is a computational strategy that examines the assortment of amino acid substitutions happening at fundamental conditions that are occurred inside of the homologous proteins of 3-D structures and convert them into virtual substitution tables. These tables have been using as a quantitative measure for predicting the structure of protein upon a change in amino acid. This server depends on the examination of ΔΔG (in kcal/mol) which gives its values of stabilizing mutations (ΔΔG > 2.5 kcal/mol) and destabilizing changes (ΔΔG < -2.5 kcal/mol).34,35 DUET is a web server utilized for applying and considering incorporated missense mutations and their impact in the structure of proteins by computational methodology. DUET blends two adjusting techniques mCSM and SDM, that combining the results of the various procedures in an enhanced marker using Support Vector Machines (SVM). mCSM relies upon graph-based technique and predicts not simply the effect of single-point changes on steadiness quality of protein, yet moreover protein-protein and protein-nucleic acid binding.36 Proteins are extremely basic and a unique particle whose natural capacity relates to atomic movements. Regardless the fundamental role of protein elements, their computational recreation cost has provoked for the most part structure-based procedures for looking at the impact of mutations on structure and function of protein relying on static structures.37 DynaMut webserver executes two discrete yet straightforward ways to deal with assessing and visualize the structure of a protein by an assortment of adaptations and evaluation of the role of mutations on dynamicity and strength of protein following in changes vibrational entropy. This server could be beneficial in two manners; one can be utilized to decide and to contemplate protein elements and the subsequent, it may be utilized to survey the effect of single point change. The cut off worth set for this instrument is ΔΔG≥0 as balancing out and ΔΔG < 0 as destabilizing.38 DynaMut gears NMA entirely through two various advances, Bio3D and ENCoM, giving quick and essential access to an understandable and keen examination of protein movements.39,40 Normal Mode Analysis (NMA) is a computational strategy that approximates the dynamic practicality of a structure around an adjustment through consonant development. This has been used to make possible improvements and appropriately give beneficial bits of information into protein developments, and their accessible conformational assortments.41
mStructural basis protein Confirmation changes
In conformational changes studies was done by DynaMut webserver where interatomic interaction studies recognize that macro or micro molecules. This interaction studies using a pairwise alignment algorithm where the outcome shows how much mutation disrupts the interacting bonds.41
Point mutation analysis in stress condition (pH variation)
Mutational analysis of the point mutation within the variation of temperature and pH (a stress condition) on the structure-based analysis was done by MAESTROweb server. MAESTRO stands for (Multi AgEnt Stability prediction) webserver which is free for non-commercial use and based on command-line program for analysing the consequences of point mutation on innumerable pH which supports for conclusion the effect of that mutation in diseases. MAESTRO webserver apparatuses a machine learning system scans for the controlled mutation, expects ΔΔG value and estimate of steadying disulfide bonds and pragmatic for the biological molecule. MAESTRO server delivers a further remuneration which defined as (i) statement of confidence value of prediction, (ii) process on multimeric proteins, (iii) a scan mode for the most (de)stabilizing point mutations and (iv) evaluation of potential disulfide bonds. A negative value of ΔΔG means an increase in the destability of the protein whereas a positive value depicts diminish in the stability of the protein.42,43
Homology modelling of SARS CoV-2 RdRp was done which showed sequence identity of 97.05% with the template PDB ID: 6NUR (SARS-NSP12) (Figure 1).15 This 6NUR structure is the complex structure of NSP12 with NSP7 and NSP8 with 3.1 Å resolution. The generated SARS CoV-2 RdRp structure QMEAN (qualitative model energy analysis) score is -0.68 which is good for further study.17 The model also includes two zinc ions and it encompasses residues 4501-5315 of the SARS CoV-2 orf1ab. Model validation by SAVES, the Verify3D server evaluated scoring 91.16% of the residues have average 3D-1D score >= 0.2 and the scoring pass while the quality factor predicted by ERRAT server is 95.9839. The Ramachandran plot is 92.5% in allowed region additional allowed region is 7.2% else generously allowed region is 0.3% and disallowed region is 0.0%.18–21
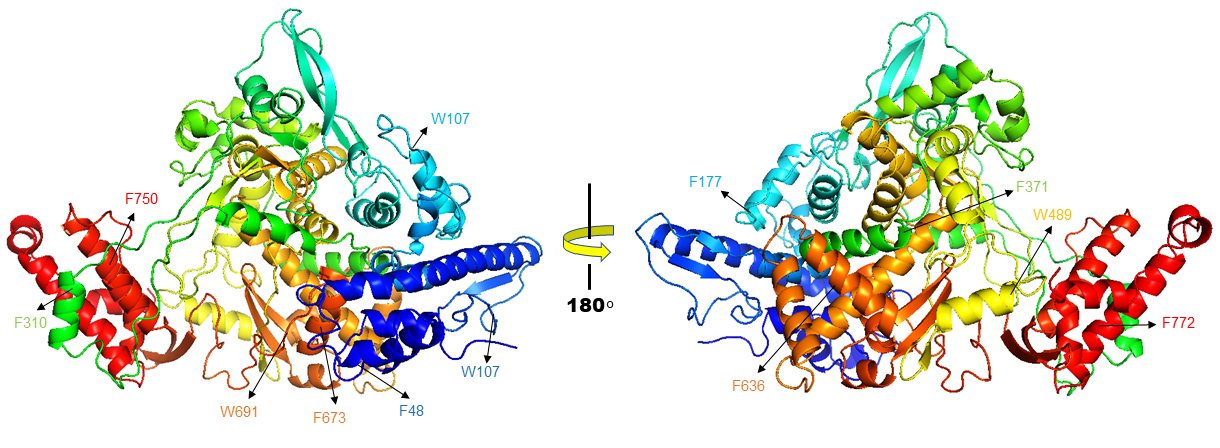
Figure 1 Cartoon representation of NSP12 from SARS CoV-2, Here predicted twelve residues (F48, W107, W159, Y177, F310, F371, W489, F636, F673, W691, F750 AND F772) which showing mutation on these residues changes the protein confirmation which results highest stability decrease of NSP12 protein.
Selected residues for calculate Structural stability of NSP12
For the selection of residues using EASE-MM server which is a sequence-based predicted mutated amino acid possibility regarding protein stability. EASE-MM server comprises of five specialized models of support vector machines and then select final prediction based on selected secondary structure model and its surface area. After EASE-MM server predicted outcome selected residues as shown in Figure 1, there were twelve different residues which values are in -5 are as F48, W107, W159, Y177, F310, F371, W489, F636, F673, W691, F750 AND F772 as shown in Table 1.23,24 This server predicted mutation of different residues into Glycine (G) outcome highest decrease stability in NSP12 protein, mutation into G has shown more negative score rather than other residues which means more stability decreases of the protein.
|
S.no. |
Mutation |
ΔΔG |
Stability class |
SS |
rASA |
|
1 |
F48G |
-5.0208 |
destabilising |
H |
0.09 |
|
2 |
W107G |
-5.1435 |
destabilising |
E |
0.22 |
|
3 |
W159G |
-5.1142 |
destabilising |
E |
0.11 |
|
4 |
Y177G |
-5.0648 |
destabilising |
H |
0.07 |
|
5 |
F310G |
-5.1467 |
destabilising |
H |
0.08 |
|
6 |
F371G |
-5.0227 |
destabilising |
H |
0.07 |
|
7 |
W489G |
-5.1482 |
destabilising |
H |
0.1 |
|
8 |
F636G |
-5.284 |
destabilising |
H |
0.03 |
|
9 |
F673G |
-5.2878 |
destabilising |
H |
0.03 |
|
10 |
W691G |
-5.2553 |
destabilising |
E |
0.23 |
|
11 |
F750G |
-5.101 |
destabilising |
H |
0.04 |
|
12 |
F772G |
-5.0087 |
destabilising |
H |
0.04 |
Table 1 EASE-MM server predicted the mutation-induced stability changes resulting in top 12 mutation hits has greater decreases stability in (-5).
SS, Secondary Structure; H, helix; E,e xtended/sheet; rASA, relative accessible surface area.
Sequence-based analysis of point mutations
PROVEAN v1.1.3: For the further assessment of mutations, we have used PROVEAN server which gives result that shows the effect of mutations on protein structure and define a mutation as deleterious or non-deleterious. PROVEAN sever predicted all the selected residues mutation cause the deleterious effect but the highest larger negative value is on W489G (-12.711) and W691G (-12.663) rest residues score are shown in Table 2.25,26
|
S.no. |
Variant |
PROVEAN score |
Deleterious (cutoff= -2.5) |
|
1 |
F48G |
-7.652 |
Deleterious |
|
2 |
W107G |
-6.27 |
Deleterious |
|
3 |
W159G |
-5.36 |
Deleterious |
|
4 |
Y177G |
-9.509 |
Deleterious |
|
5 |
F310G |
-8.692 |
Deleterious |
|
6 |
F371G |
-7.692 |
Deleterious |
|
7 |
W489G |
-12.711 |
Deleterious |
|
8 |
F636G |
-7.278 |
Deleterious |
|
9 |
F673G |
-8.767 |
Deleterious |
|
10 |
W691G |
-12.663 |
Deleterious |
|
11 |
F750G |
-6.458 |
Deleterious |
|
12 |
F772G |
-8.555 |
Deleterious |
Table 2 PROVEAN server predicted selected mutate residue PROVEAN score which shows deleterious effect (cut off= -2.5)
F, Phenylalanine; W, Tryptophan; Y, Tyrosine; G, Glycine.
iStable server: NSP12 protein sequence-based analysis of the various mutant residues was done by iStable server whose predicted result shows the wild type form to mutant form. Therefore, all the twelve selective residues (F48, W107, W159, Y177, F310, F371, W489, F636, F673, W691, F750 AND F772) has decreases the stability of the protein whose result is shown in Table 3.28,29
|
S.No. |
Mutation |
iMutant2.0SEQ/ ΔΔG |
MUpro/Cnf. Score |
iStable/Cnf. Score |
|
1 |
F48G |
Decreases/ -2.50 |
Decreases/ -1 |
Decreases/ 0.8651 |
|
2 |
W107G |
Decreases/ -2.22 |
Decreases/ -0.89 |
Decreases/ 0.8000 |
|
3 |
W159G |
Decreases/ -2.11 |
Decreases/ -1 |
Decreases/ 0.8625 |
|
4 |
Y177G |
Decreases/ -2.96 |
Decreases/ -1 |
Decreases/ 0.8122 |
|
5 |
F310G |
Decreases/ -2.81 |
Decreases/ -1 |
Decreases/ 0.8430 |
|
6 |
F371G |
Decreases/ -2.56 |
Decreases/ -1 |
Decreases/ 0.8945 |
|
7 |
W489G |
Decreases/ -2.68 |
Decreases/ -0.95 |
Decreases/ 0.7957 |
|
8 |
F636G |
Decreases/ -3.45 |
Decreases/ -1 |
Decreases/ 0.8818 |
|
9 |
F673G |
Decreases/ -2.78 |
Decreases/ -1 |
Decreases/ 0.8903 |
|
10 |
W691G |
Decreases/ -2.45 |
Decreases/ -1 |
Decreases/ 0.8449 |
|
11 |
F750G |
Decreases/ -3.04 |
Decreases/ -1 |
Decreases/ 0.8674 |
|
12 |
F772G |
Decreases/ -2.93 |
Decreases/ -1 |
Decreases/ 0.9106 |
Table 3 iStable metaserver sequence-based mutation predicted (ΔΔG) decrease stability changes
F, Phenylalanine; W, Tryptophan; Y, Tyrosine; G, Glycine.
STRUM (mutation stability changes prediction): STRUM server based on Pearson correlation coefficient (PCC) between predicted and measured changes of the Gibbs free-energy gap. STRUM had been utilized to investigate the prospect for improving ΔΔG prediction based on low-resolution models from the iterative assembly refinement (I-TASSER) simulations, in which three groups of features from sequence outline, multiple template threading, and I-TASSER atomic models were combined. STRUM server results show W107, W159, Y177 and W691 have negative values which means a decrease stability changing on sequence basis point mutation rest followed residues computed protein stability changes upon point mutation as shown in Table 4.32,33
|
S.No. |
Position |
Wild type |
Mutant type |
Sequence basis ∆∆G |
|
1 |
48 |
F |
G |
0.14 |
|
2 |
107 |
W |
G |
-0.2 |
|
3 |
159 |
W |
G |
-1.18 |
|
4 |
177 |
Y |
G |
-0.26 |
|
5 |
310 |
F |
G |
0.77 |
|
6 |
371 |
F |
G |
0.45 |
|
7 |
489 |
W |
G |
0.06 |
|
8 |
636 |
F |
G |
1 |
|
9 |
673 |
F |
G |
1.3 |
|
10 |
691 |
W |
G |
-1.12 |
|
11 |
750 |
F |
G |
1.03 |
|
12 |
772 |
F |
G |
0.89 |
Table 4 Point mutation analysis by using STRUM server
F, Phenylalanine; W, Tryptophan; Y, Tyrosine; G, Glycine.
Structure-based analysis of point mutations
DUET server: In this server, there are two other approaches functional (mCSM and SDM) in a consent keenness, usual by combining the consequences of the different approaches in an improved pointer applying SVM methods. DUET server predicted the single residue point mutation which computes the structural stability. In these studies, the outcome of mCSM, SDM and DUET calculates ΔΔG value which predicted protein destabilizing here the output data shows all the selected point residues for mutation has negative values means it confirms every point mutations has decreases the protein destability whose values had been shown in Table 5.36
|
S.No. |
Wild type |
Residue position |
Mutant type |
mCSM predicted ∆∆G |
SDM predicted ∆∆G |
DUET predicted ∆∆G |
|
1 |
F |
48 |
G |
-2.776 Kcal/mol |
-2.22 Kcal/mol |
-3.064 Kcal/mol |
|
2 |
W |
107 |
G |
-2.645 Kcal/mol |
-1.83 Kcal/mol |
-2.466 Kcal/mol |
|
3 |
W |
159 |
G |
2.596 Kcal/mol |
0.06 Kcal/mol |
-2.075 Kcal/mol |
|
4 |
Y |
177 |
G |
-2.908 Kcal/mol |
-1.99 Kcal/mol |
-3.119 Kcal/mol |
|
5 |
F |
310 |
G |
-2.166 Kcal/mol |
-2.33 Kcal/mol |
-2.438 Kcal/mol |
|
6 |
F |
371 |
G |
-3.205 Kcal/mol |
-2.41 Kcal/mol |
-3.528 Kcal/mol |
|
7 |
W |
489 |
G |
-3.864 Kcal/mol |
-2.53 Kcal/mol |
-3.522 Kcal/mol |
|
8 |
F |
636 |
G |
-2.158 Kcal/mol |
-2.33 Kcal/mol |
-2.429 Kcal/mol |
|
9 |
F |
673 |
G |
-3.55 Kcal/mol |
-2.33 Kcal/mol |
-3.795 Kcal/mol |
|
10 |
W |
691 |
G |
-3.444 Kcal/mol |
-0.06 Kcal/mol |
-2.859 Kcal/mol |
|
11 |
F |
750 |
G |
-2.419 Kcal/mol |
-2.33 Kcal/mol |
-2.697 Kcal/mol |
|
12 |
F |
772 |
G |
-3.956 Kcal/mol |
-2.33 Kcal/mol |
-4.154 Kcal/mol |
Table 5 DUET server predicted protein structure stability (∆∆G) comparative studies between mCSM and SDM servers
F, Phenylalanine; W, Tryptophan; Y, Tyrosine; G, Glycine.
SDM server: Structure-based point mutation calculating score prediction Site Directed Mutator (SDM server) computes the comprises study of wild-type and mutated secondary structure evaluation, solvent accessibility, depth (Å) of the protein and ∆∆G value which predicted protein destability. For calculating protein stability changes using thermodynamic cycle (∆GFwt to mt). SDM computed scores (∆∆G) showing except W159 and W691 all the remaining mutated residues has large decrease protein stability as shown in Table 6.35 The resulting outcome mostly residues have negative value which predicted that may be mutation on these residues of a point caused protein malfunction.
|
S.No. |
Position |
Predicted ∆∆G |
Wild type |
Mutant type |
||||
|
Secondary structure |
S A % |
Depth (Å) |
Secondary structure |
S A % |
Depth (Å) |
|||
|
1 |
F48G |
-2.04 |
H |
9.4 |
5.2 |
H |
39.6 |
4.1 |
|
2 |
W107G |
-1.77 |
b |
4.5 |
6.4 |
b |
31.3 |
3.9 |
|
3 |
W159G |
0.23 |
p |
20.2 |
4.5 |
p |
58.8 |
3.6 |
|
4 |
Y177G |
-3.17 |
H |
0 |
6.9 |
H |
22.6 |
7 |
|
5 |
F310G |
-2.78 |
H |
0 |
9.3 |
H |
8.6 |
6.8 |
|
6 |
F371G |
-2.14 |
a |
4.3 |
9.5 |
a |
54.6 |
5.9 |
|
7 |
W489G |
-3.37 |
H |
2.4 |
7.1 |
H |
29.2 |
6.7 |
|
8 |
F636G |
-2.78 |
H |
0.6 |
10 |
H |
25.3 |
6.3 |
|
9 |
F673G |
-2.78 |
H |
0 |
10.3 |
H |
52.1 |
9 |
|
10 |
W691G |
0.11 |
b |
17.9 |
5.5 |
b |
61.6 |
4.2 |
|
11 |
F750G |
-2.78 |
H |
2.4 |
8.5 |
H |
26.8 |
6.8 |
|
12 |
F772G |
-2.78 |
H |
0.1 |
6.7 |
H |
8.2 |
7 |
Table 6 SDM server predicted the structure stability ∆∆G and topological properties in between wild type and mutant (Secondary Structure, Solvent accessibility and Depth)
H-helix; a, p, b-α-helix and β-sheets classified according to their mainchain φ-ψ torsion angles; SA%, Solvent accessibility.
DynaMut server: Impact changes on point mutation by DynaMut server used two diverse ways, (1) study of protein dynamics and (2) effect of point mutation on protein structural stability. Here using NSP12 protein PDB file studied point mutation effect on protein flexibility and structural destabilizing effect. In this server, mutation study includes three different servers mCSM, SDM and DUET which was calculated earlier in this manuscript.38 Here normal mode analysis also shows the destabilizing effect of the mutation by two different approaches Bio3D and ENCoM. ENCoM server also computes the protein structure ∆∆Svib vibrational entropy between wild-type and mutant.
DynaMut server predicted all the selected residues for point mutation has a negative value which shows the destabilizing effect whose values had been shown in (Table 7–9).39,40 The vibrational entropy energy between wildtype and mutant of all twelve residues are F48, W107, W159, Y177, F310, F371, W489, F636, F673, W691, F750 AND F772 which are showing mutated residues increase of molecule flexibility as shown in Figure 2.41
|
S.No. |
Wild type |
Residue position |
Mutant type |
DynaMut predicted ∆∆G |
NMA based ∆∆G ENCoM |
∆∆Svib ENCoM |
|
1 |
F |
48 |
G |
-2.847 kcal/mol |
-0.978 kcal/mol |
1.223 kcal.mol-1.K-1 |
|
2 |
W |
107 |
G |
-1.805 kcal/mol |
-1.633 kcal/mol |
2.041 kcal.mol-1.K-1 |
|
3 |
W |
159 |
G |
-1.605 kcal/mol |
-1.234 kcal/mol |
1.543 kcal.mol-1.K-1 |
|
4 |
Y |
177 |
G |
-2.453 kcal/mol |
-1.361 kcal/mol |
1.701 kcal.mol-1.K-1 |
|
5 |
F |
310 |
G |
-0.662 kcal/mol |
-1.575 kcal/mol |
1.969 kcal.mol-1.K-1 |
|
6 |
F |
371 |
G |
-3.340 kcal/mol |
-1.092 kcal/mol |
1.365 kcal.mol-1.K-1 |
|
7 |
W |
489 |
G |
-2.940 kcal/mol |
-1.730 kcal/mol |
2.162 kcal.mol-1.K-1 |
|
8 |
F |
636 |
G |
-1.709 kcal/mol |
-1.141 kcal/mol |
1.426 kcal.mol-1.K-1 |
|
9 |
F |
673 |
G |
-3.324 kcal/mol |
-1.475 kcal/mol |
1.844 kcal.mol-1.K-1 |
|
10 |
W |
691 |
G |
-1.780 kcal/mol |
-1.405 kcal/mol |
1.756 kcal.mol-1.K-1 |
|
11 |
F |
750 |
G |
-1.414 kcal/mol |
-1.516 kcal/mol |
1.895 kcal.mol-1.K-1 |
|
12 |
F |
772 |
G |
-2.655 kcal/mol |
-1.591 kcal/mol |
1.988 kcal.mol-1. K-1 |
Table 7 DynaMut server predicted structural stability ∆∆G (kcal/mol) and vibrational entropy energy between wild type and mutant ∆∆Svib (kcal.mol-1. K-1)
|
S.No. |
Residue position |
pH variation (stress conditions) |
||||||
|
5.5 |
6 |
6.5 |
7 |
7.5 |
8 |
8.5 |
||
|
1 |
F48G |
-0.16 |
-0.047 |
0.219 |
0.349 |
0.391 |
0.392 |
0.365 |
|
2 |
W107G |
0.04 |
0.126 |
0.355 |
0.704 |
0.842 |
1.141 |
1.262 |
|
3 |
W159G |
0.13 |
0.195 |
0.218 |
0.309 |
0.508 |
0.63 |
0.879 |
|
4 |
Y177G |
0.08 |
0.291 |
0.366 |
0.7 |
0.833 |
0.959 |
1.061 |
|
5 |
F310G |
0.05 |
0.235 |
0.373 |
0.615 |
0.73 |
0.804 |
0.952 |
|
6 |
F371G |
-0 |
0.19 |
0.331 |
0.613 |
0.697 |
0.701 |
0.884 |
|
7 |
W489G |
0.18 |
0.251 |
0.368 |
0.635 |
0.869 |
1.047 |
1.081 |
|
8 |
F636G |
0.06 |
0.271 |
0.383 |
0.767 |
0.757 |
0.866 |
1.007 |
|
9 |
F673G |
-0.03 |
0.195 |
0.244 |
0.527 |
0.659 |
0.737 |
0.916 |
|
10 |
W691G |
0.24 |
0.235 |
0.341 |
0.298 |
0.596 |
0.583 |
0.652 |
|
11 |
F750G |
0.06 |
0.235 |
0.369 |
0.652 |
0.719 |
0.822 |
0.97 |
|
12 |
F772G |
0.07 |
0.248 |
0.383 |
0.679 |
0.752 |
0.855 |
0.997 |
Table 8 MAESTROweb predicted structure stability (∆∆G) on different pH variation
|
S.No. |
Web server |
Input method |
Used algorithm |
URL |
Ref. |
|
1 |
Ease-MM |
Protein sequence |
Using support vector machine (SVM) |
http://sparks-lab.org/server/ease/ |
23 |
|
2 |
PROVEAN |
Protein sequence |
It searched against the NCBI nr database using BLAST and clustered using CD-HIT |
http://provean.jcvi.org/index.php |
26 |
|
3 |
iStable |
Protein sequence |
Using support vector machine (SVM) |
http://predictor.nchu.edu.tw/istable/ |
28 |
|
4 |
I-Mutant2.0 |
Protein sequence |
SVM regression with a radial basis function kernel, and RSA |
http://gpcr2.biocomp.unibo.it/cgi/predictors/I-Mutant3.0/I-Mutant3.0.cgi |
30 |
|
5 |
MUpro |
Protein sequence |
Using Feed-Forward Neural Network and SVMs |
http://mupro.proteomics.ics.uci.edu/ |
31 |
|
6 |
STRUM |
Protein sequence |
Pearson correlation coefficient (PCC) between predicted and measured changes of Gibbs free-energy gap |
https://zhanglab.ccmb.med.umich.edu/STRUM/ |
32 |
|
7 |
SDM |
Protein structure |
Environment-specific substitution tables (ESSTs) |
http://marid.bioc.cam.ac.uk/sdm2 |
35 |
|
8 |
DUET |
Protein structure |
SVM regression with a Radial Basis Function kernel and RSA |
http://biosig.unimelb.edu.au/duet/ |
36 |
|
9 |
DynaMut |
Protein structure |
Graph-based signatures in a consensus predictor |
http://biosig.unimelb.edu.au/dynamut/ |
38 |
|
10 |
MAESTROweb |
Protein structure |
As a Command line web user interface |
https://biwww.che.sbg.ac.at/maestro/web |
42,43 |
Table 9 In this table enlists the computational online server used in this study
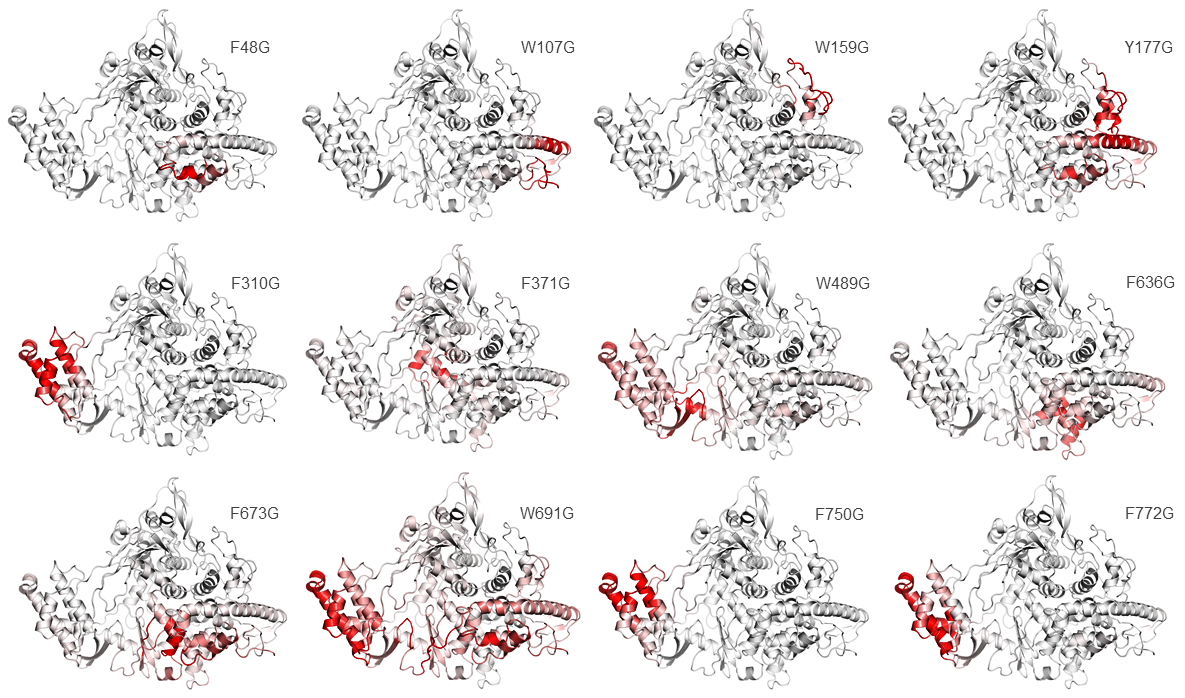
Figure 2 Effect of Mutated residues predicted the molecule flexibility via Vibrational entropy energy. In figure showing (red color) predicted mutated residues changes the protein confirmation which increases the molecule flexibility.
Point mutation calculated Interatomic interaction changes
The essential studies of point mutation are the analysis of interatomic changes into wildtype to a mutant which was done by DynaMut web server it allows the visualization of all interactions which are calculated by Arpeggio web server. This server also computes the deformation energies and molecular and atomic fluctuations between wild type and mutant protein PDB structures. The point mutation of all sites which are described earlier in results, visualization of the disruption bond like hydrogen bonds, halogen bonds and hydrophobic between different atoms of the protein. In interatomic interaction studies, the mutant W107, W159 and F636 with Glycine showing a highly drastic changes and other mutation residues also have shown bond disruption showing in Figure 3.33,41
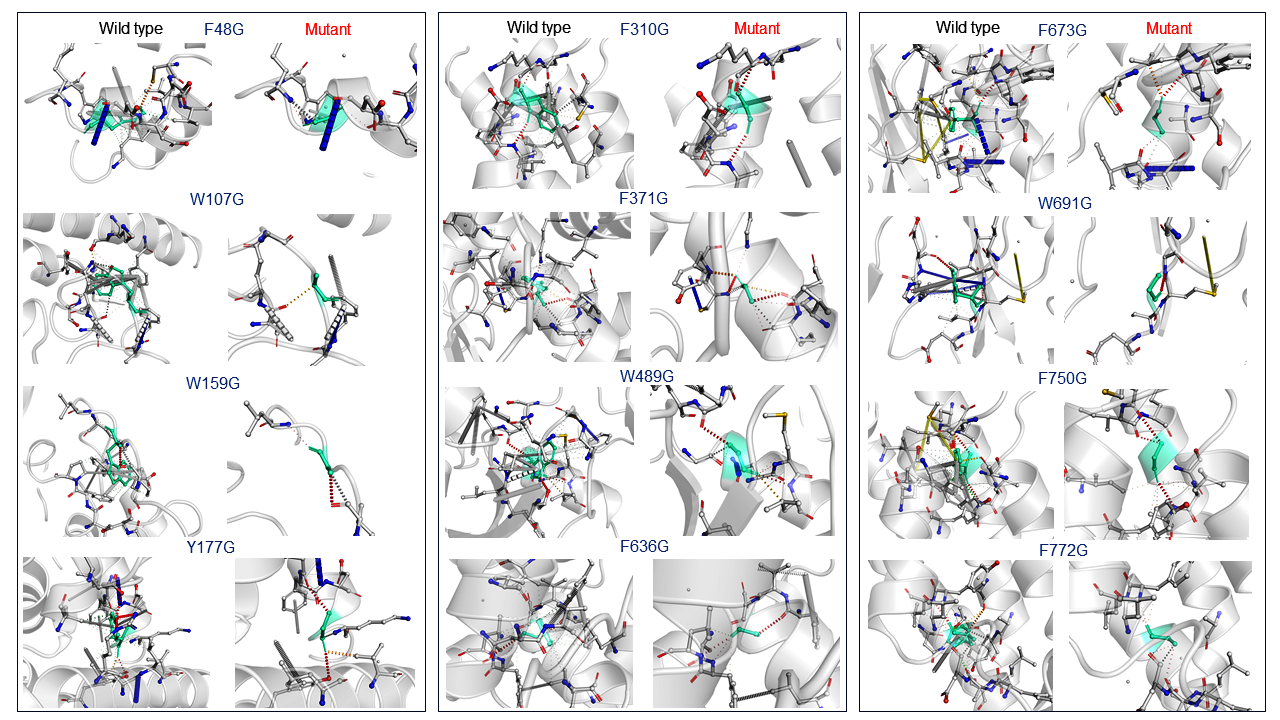
Figure 3 In this figure showing the interatomic interaction between wildtype and mutant which shows in twelve residues there is three mutant W107, W159 and F636 with Glycine showing a highly drastic changes and other mutation residues also have shown bond disruptions.
Point mutation analysis in Stress Condition (pH variation)
MAESTROweb server used for the further studies of the effect of different pH of point mutation. This server as a command-line web user interface which input is structure basis. Structure-based point mutation analysis with various pH (5.5, 6, 6.5, 7, 7.5, 8, and 8.5) was done by MAESTROweb server. This program is based on machine learning method and predicts the connotation of single amino acid mutation on (5.5 to 8.5) range of pH and their negatives values means an increase in the protein stability whereas positive values vice versa. This server result shows that pH 8.5 had greater decreases in protein stability except F48G had greater decreases in protein stability on pH 7.5-8 which is shown in Figure 4.42,43
COVID-19 outbreak after in Wuhan city occurs in the whole world without having any permanent cure for this. This is the third consecutive occurrence of SARS virus after SARS and MERS virus outbreaks in 2003 and 2014 respectively that runs almost 10 million people to the hospital and severely affected the economy of each country. NSP12 is an essential part of NSP series protein and important for RNA dependent RNA polymerase activity. In this study the identification and understanding the role of point mutation on the structural stability of NSP12. In this report, by using computational approaches predicted the deleterious effect of selected point mutation on structural stability. Similarly, the online server EASE-MM, PROVEAN, iSTABLE, STRUM, DUET, SDM and DynaMut showing decrease in protein stability effect. In interatomic interaction analysis the mutant variant W107, W159 and F636 with Glycine showing a highly deleterious effect on the structural stability of NSP12 and in depth this point mutations studies specifying the drastic changes in NSP12 structural stability. This study proposed a full vision to understand the molecular basis of NSP12 which may be utilized in targeting this gene for therapeutic strategies.
The authors acknowledge support from the Centre for Interdisciplinary Research in Basic Science, Jamia Millia Islamia University. Md Amjad Beg also acknowledges UGC-MANF (MANF-2017-18-UTT-88071) for the financial support and Jamia Millia Islamia University.
The authors declare that they have no potential conflict of interests.
Md Amjad Beg: Conceptualization idea, Methodology designing, Software handling, writing manuscript and evaluate results.
Dr Fareeda Athar: Reviewing and corresponding author.

©2020 Beg, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.