Journal of
eISSN: 2373-6453


Research Article Volume 1 Issue 3
1Department of Animal Resources, Qatar
2Department of Virology, Cairo University, Egypt
Correspondence: M Haroun, Virology Unit, Veterinary Laboratory, Department of Animal Resources, 23211, Doha, Qatar, Tel +974-44560-546, Fax +974-44653-086
Received: June 30, 2014 | Published: July 22, 2014
Citation: Abdulla NM, Haroun M, Shalaby MA, Elsanousi AA (2014) Comparative Study on Some Characteristics of Newcastle Disease Virus Field Strains Isolated From Captivated Avian Species in Qatar. J Hum Virol Retrovirol 1(3): 00014. DOI: 10.15406/jhvrv.2014.01.00014
Six Newcastle disease virus (NDV) field strains (NDV-QF10, NDV-QF13, NDV-QH10-1, NDV-QH10-2, NDV-QQ09 and NDV-QP09) were isolated and pathotyped from samples collected from captivated falcons, houbara bustard, quail and pigeon, respectively, during the study period of 2012 and 2013 in Qatar. Virus cultivation in specific pathogen free embryonating chicken eggs (SPF ECE), haemagglutination (HA), haemagglutination inhibition (HI) and mean death time (MDT) assays used in the investigation displayed virus identity and pathogenicity. The six NDV isolates were classified velogenicity strains. Sequence analysis of 150bp of the amplified 216bp nucleotides of the functional region of the F-gene encoding the F2-F1 cleavage site, and phylogenies of three of the six strains revealed 112R-R-Q-K-R-F117 amino acid motif confirming velogenicity, and that, the three strains clustered under NDV genotype VII either subgroup d or f.
Keywords: Newcastle disease virus, Captivated avian species, Virus isolation, Pathotyping, Phylogenetic analysis, Genotyping
ND, Newcastle Disease; NDV, Newcastle Disease Virus; SPF-ECE, Specific Pathogen-Free Embryonating Chicken Eggs; AAF, Amnio Allantoic Fluid; HA, Haemagglutination; HAU, Haemagglutinating Units; HI, Haemagglutination Inhibition; MDT, Mean Death Time; ICPI, Intracerebral Pathogenicity Index; RT-PCR, Reverse Transcription Polymerase Chain Reaction; cDNA, complementary Deoxynucleotide Acid
Newcastle disease (ND) is a known highly contagious disease of a significant worldwide impact on avid species. The virus belongs to avian paramyxovirus type 1(APMV-1) serotype, genus Avulavirus that relates to the family Paramyxoviridae and Mononegavirales order.1,2 Hundreds species were estimated to be susceptible3 rendering serious losses among poultry rearing and industry economy. Beside, the virus possess zoonotic characteristics that causing mild conjunctivitis to mankind. Since the first informative detection, isolation and conventional pathoyping of Newcastle disease virus (NDV) in Qatar,4 continuous reporting of the disease was reported. Focal annual outbreaks of 120, primarily among chickens, pigeons and other domesticated birds were observed.5 This situation signals public health hazards to tendency to rear feral birds, the estimated 3.5 million traditional poultry system that represents 23% of the total bird population, and the 11 million bird of the growing industrial poultry business that valued to 77% of the state poultry population.6
Aiming to have a preliminary insight on the nature of Newcastle disease virus (NDV) strains circulating in captivated avid species reared in Qatar, conventional virological methods coupled with late molecular assays were used to reveal some properties of the strains including virulence, genotype and phylogenic relatedness to other known worldwide NDV strains.
Samples
Trachea land cloacal swabs, spleen and liver samples collected from 51 apparently healthy and ND-suspected captivated falcons (Falco peregrines peregrinus), diseased Houbara (Chlamydotis undulata), quail (Colinus virgin/anus) and pigeons (Columba livia) were used for the study. Swabs were immediately transported in cool chamber condition and frozen at -40oC till used. Positive amnio allantoic fluids (AAF) from specific pathogen free (SPF) embryonating chicken eggs (ECE) inoculated with the collected samples were either used freshly or as frozen stocks.
Virus isolation
Tracheal and cloacal swabs ‘elution and tissue homogenates were used or virus isolation. Ten per cent of each sample was prepared in sterile phosphate-buffered saline (SPBS), followed by centrifugation for 10 min at 2500 rpm at 20oC. Each of the final inoculum was supplemented with 100 μL 200 IU mL-1 cell culture grade penicillin, 100 mg mL-1 streptomycin and 20 mg mL-1 gentamycin suspensions. Two hundred μL of each swab elution and tissue homogenate was inoculated into the allantoic sac of five 10-day-old SPF ECE, Loghman breed (VALO, Germany). Controls received the same dose of SPBS. All eggs were incubated at 37oC for 8 days observation. Two hundred microlitre of each neat AAF obtained from the first passage was inoculated for a successive second passage using the same inoculation conditions.
Haemagglutiantion (HA) and Haemagglutination Inhibition (HI) test
AAF harvest of passage level two was used for virus identification applying HA and HI assay following standard procedures.7 Ten percent 4-week-old washed chicken RBC was used for rapid spot HA testing individual AAF of each dead embryo prior to harvesting and pooling. Four HAU determined by HA assay of the positive AAF pools of each isolate was used in HI to determine the virus strains specificity to the reference anti-NDV serum (PA0155, VLA, UK).
Determination of the 50 Embryo Lethal Dose (ELD50), Minimum Embryo Lethal Dose (MELD) and Mean Death Time (MDT)
MDT determination follows8 standard procedure with minor modifications. A single-step determination of ELD50, MELD and MDT was conducted. Two hundred microlitre of 105-1015 AAF dilutions prepared from the second passage was inoculated each into the allantoic sac of five10-day-old SPF ECE (Loghman breed; VALO, Germany). All eggs including uninfected controls were incubated at 37oC and observed for 8 days at 12 h intervals. ELD50 was calculated according to.9
Viral RNA extraction
Pools of positive haemagglutination (HA)/haemagglutination inhibition (HI) NDV AAF were used for viral RNA extraction using QIAamp (QIAamp ViralRNA Mini Handbook, 12/2005). Briefly, 140µL of each pool was added to 560µL buffer AVL RNA carrier, pulse-vortexed for 15 sec and incubated at room temperature (15-25oC) for 10 min. 560µL 96-100% ethanol was used for precipitation purposes. QIAamp Mini spin column contained in a 2 mL collection tube was used for loading of the lysed material. 500µL Buffer AWI and AW2 were used successively to maintain RNA binding and washing. Finally, 80µL AVE Buffer was used to elude the RNA extract after 1 min room temperature incubation and 1 min at 6000X g (8000rpm) centrifugation. All viral RNA extracts were stored at -20oC till amplification.
Reverse transcription polymerase chain reaction (RT-PCR)
The forward (5’- GCA GCTGCA GGG ATT GTG GTG-3’, nucleotide position 158-177) and reverse (5’- TCT TTG AGCAGG AGG ATG TTG-3’, nucleotide position 513-493) oligo nucleotide primer sets were used for amplification of 356b pamplicons corresponding the cleavage activation site of NDV-F gene.10,11 RT-PCR amplification followed Access Quick RT-PCR Kit (Cat. no. A1700). 25.5µL total reaction mix was prepared using 12.5µL 2X Access Quick Master Mix, 2.5µL 1 µM upstream F-primer and control R-primer each, 2.5µL Nuclease-free water, 0.5 µL 5 U/µ AMV RT enzyme. A three-step amplification method of 35-cycle-programmed PCR machine (9800 Fast Thermal Cycler) was used for cDNA amplification as follows: 45oC for 45 min and 95oC for 2 min (RT-step), 3-step cycling of 94oC for 45 sec (denaturation), 58oC for 45 sec(annealing), 72oC for 45 sec (extension) and 72oC for 5 min (final extension).
Sequence analysis of the complementary deoxynucleotideacid (cDNA) fragments
cDNART-PCR products were measured for concentration and run on 1.5% agarose gel according to standard procedures. Fragments’ purification was done according to ExoSAP-IT instructions. Amplification was performed with the 216bp NDV F-gene nested primer sets (F-primer: 5′-CCC CGT TGGAGG CAT AC-3′, nucleotide positions 282-298; R-primer: 5′-TGT TGG CAG CAT TTT GATTG-3′, nucleotide positions 497-478) according to ExoSAP-IT instructions (PCR: 78200/01/02/05/50-Affymetrix, USA) using 16.2μL ddH2O, 0.8μL each F and R primers, 1μL purified cDNA product. The deduced nucleotide sequences were generated constructed and aligned using MEGA (Version 5).
The epidemiological and clinical findings of this investigation demonstrated that all of the birds’ populations involved in these focal outbreaks were susceptible to NDV and not vaccinated. ND-infected birds demonstrated a rapid disease course pattern of about 1-3 days. All birds showed nervous signs. Beside, the houbara and quails showed diarrhea. The quail necropsies exhibited ecchymosis in the mucosa of the proven triculus and intestines. Based on successful recovery of the strains on inoculation of the suspected NDV swabs’ elution samples and tissue homogenates into AAF of the 10-day-old SPF ECE, exhibition of the biological HA characteristics, HI of the isolates on testing against the reference anti-NDV serum with a minimum HI values of 4log2 (Table 1), retrieval of the specific 356bp NDV F-gene products at the cleavage site of the NDV F0 protein using RT-PCR; six NDV field strains were isolated.
NDV Strain |
Species |
HA (Log 2) |
HI (> 4Log 2) |
ELD50 (Log 10) (1mL) |
MELD (Log 10) (1mL) |
MDT (Hour) |
ICPI |
NDV-QF10 |
Falcon |
10 |
Positive |
9.8 |
5 |
48 |
ND |
NDV-QF13 |
Falcon |
8 |
Positive |
9.7 |
6 |
48 |
ND |
NDV-QH10-1 |
Houbara |
6 |
Positive |
12.3 |
11 |
48 |
ND |
NDV-QH10-2 |
Houbara |
6 |
Positive |
9.8 |
8 |
48 |
ND |
NDV-QQ09 |
Quail |
7 |
Positive |
10.2 |
7 |
48 |
ND |
NDV-QP09 |
Pigeon |
6 |
Positive |
9.8 |
6 |
48 |
ND |
NDV-QC08 |
Chicken |
6 |
Positive |
8.2 |
7 |
60 |
1.58 |
Table 1 Some properties of the isolated NDV field strains tested at egg passage level 2 and compared to NDV-QC08 (the first NDV isolate derived from chicken in Qatar4
The fact that the isolated strains are originating from samples collected from four different avian type species (falcon, houbara, quail and pigeon) signifies the circulation of ND among Qatari birds. Further, it also indicates the endemic situation of the disease in the country. However, not necessary genetically related, the close range of the ELD50 of the isolated strains (108.8 to 1012.3 ml-1) and the similarity of the MDT (48h) indicate the close pathogenic properties among the isolated NDV strains. MDT records classify the strains pathologically velogenic. Further, the sequence analysis of the corresponding 216bp nested NDV F-gene segments of NDV-QF13, NDV-QH10-2 and NDV-QQ09 strains Figure 1 showed 112R-R-Q-K-R-F117 amino acid sequence motif indicative of velogenicity of these isolates. These findings simulate that of the first observations of4 when a velogenic NDV strain (NDV-QC08) was isolated from ND-suspected backyard chickens using ICPI. Although the investigation did not include viral tropism, the necropsy findings of 2 of the houbara and the quail were typical viscerotropic.12,13

Figure 1 Sequence analysis of the amplified F gene of NDV-QF13, NDV-QH10-2 and NDV-QQ09 showing the gene cleavage site and its deduced amino acid sequences compared to NDV-QC08 and other Genbank worldwide strains (MEGA, Tamura-Nei model 1993).
Despite all strains were successfully propagated at passage level one and two; only at dilution 108 could the study retain the biological HA ability of the NDV-QH10-1 isolate with the highest scored ELD50 value of 1012.3. This finding might further reinforce the virulence potentiality of this isolate.
Even so, the study did not apply Intracerebral pathogenic index (ICPI), a virological candidate parameter for pathotyping,14 the successful use of part of the F-gene in sequence analysis and phylogenic studies for three isolated NDV strains, reveal convenience of this assay for basic molecular pathotype characterization, confirmatory purposes and epidemiological relatedness studies.15,16
In the present investigation, it was clear that none of NDV-QF13, NDV-QH10-2 or NDV-QQ09 has 100% nucleotide homology with the GenBank-derived NDV vaccines; Hitchner B1-47, Lasota, VGGA-87 andClone30 and that they were clustered under NDV genotype VII group whereas the vaccines are under genotype II (Figure 2). These facts reflect the basic finding that the isolated field strains are of unique virulence to that of these known vaccines. The falcon (NDV-QF13) and the quail (NDV-QQ09) strains showed 100% homology in their nucleotide sequence (Figure 1 and 3), and that they were within the subgroup VII-f (Figure 2). However, both were different in nucleotide sequence correlated to NDV-QC08,17 the first known NDV field isolated in Qatar.4 Although the nucleotide sequence and homology of the houbara isolate (NDV-QH10-2) was different from that of NDV-QC08 (Figure 1 and 3), both were within the subgroup VIId (Figure 2), which was known to be responsible for most of the global ND outbreaks since the 20th century. Regional wise, NDV-QF13, the Qatari falcon strain, was found to be genetically different from the United Arab Emirate NDV field isolates (falcon strain AV 1196/96 18, genotype IVa, Genbank accession No. AY 175738, peregrine falcon strain AV 1573/92 804/90, genotype IV-c, Genbank accession no. AY175677, domesticated fowl strain AV 51/9640-1084-95, genotype IV-c, Genbank accession no. AY 175663 and ostrich strain AV1009/98 169.1668-98, genotype IVc, Genbank accession no. AY 175666). Conclusively, all these findings might reflect the variation in the epidemiology of the field NDV strains circulating in Qatar than that of the regional reference Genbank-derived strains used in the study. However, further molecular investigations with a wide-based alignment to known GenBank-derived NDV strains are recommended to have more perception about the molecular epidemiology of the Qatari NDV field strains (Table 2).
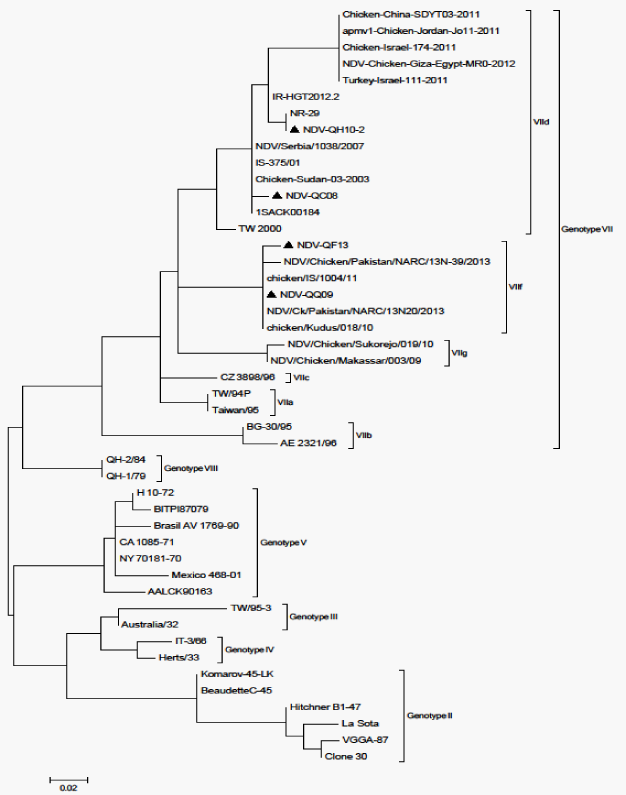
Figure 2 The phylogenetic tree analysis (MEGA) of NDV-QH10-2, NDV-QF13 andNDV-QQ09 compared to NDV-QC08 and other NDV sequences retrieved from the Genbank (Modern Evolutionary Genetic Analysis “MEGA”), Tamura-Nei model 1993).
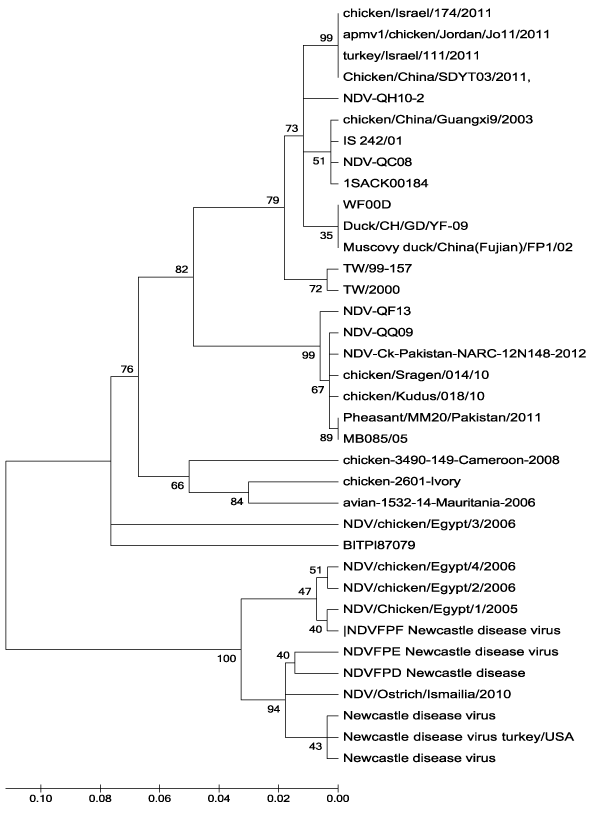
Figure 3 Homology percentage of the nucleotide sequence analysis (MEGA) of NDV-QH10-2, NDV-QF13 and NDV-QQ09 compared to NDV-QC08 and other NDV sequences retrieved from the Genbank (Modern Evolutionary Genetic Analysis “MEGA”), Tamura-Nei model 1993).
Isolate |
Host |
Protein/Gene |
Accession No. |
NDV isolate IS/375/01 |
- |
Fusion protein (F) gene- partial cds |
JF795626.1 |
NDV isolate IS/374/01 |
- |
Fusion protein (F) gene- partial cds |
JF795622.1 |
NDV isolate IS/242/01 |
- |
Fusion protein (F) gene- partial cds |
JF795621.1 |
NDV isolate Chicken/China/Guangxi9/2003 |
Chicken |
Complete genome |
JF343539.1 |
NDV isolate Chicken/China/Guangxi9/2003 |
Chicken |
Complete genome |
DQ485230.1 |
NDV isolate Duck/H/GD/YF-09 |
Duck |
Complete cds |
KC750156.1 |
NDV isolate Chicken/China/Hebei/01/2006 |
Chicken |
Complete genome |
KC542895.1 |
NDV strain WF00D |
- |
Complete genome |
FJ754272.1 |
NDV strain IR-HGT2012.2 |
- |
Fusion protein (F) gene- partial cds>JX131350.1 |
|
NDV strain IR-Ostrich-HGT2012 |
Ostrich |
Fusion (F) gene- partial cds |
JX131358.1 |
NDV isolate NR-29 |
- |
Fusion (F) gene- partial cds |
KC161990 |
NDV strain IR-HGT2010.2 |
|
Fusion protein (F) gene- partial cds |
JX131353.1 |
NDV isolate NR_91 |
|
Fusion (F) gene- partial cds |
JX129804.1 |
NDV strain IR-HGT2012.2 |
- |
Fusion protein (F) gene- partial cds>JX131350.1 |
JX131358.1 |
NDV strain IR-Ostrich-HGT2012 |
Ostrich |
Fusion protein (F) gene- partial cds |
|
NDV strain IR-HGT2012.1 |
- |
Fusion (F) gene- partial cds |
JX131357.1 |
NDV isolate NR_98 |
- |
Fusion (F) gene- partial cds |
JX129809.1 |
NDV isolate NR_97 |
- |
Fusion (F) gene- partial cds |
JX129808.1 |
NDV isolate NR_95 |
- |
Fusion (F) gene- partial cds |
JX129807.1 |
NDV isolate NR_94 |
- |
Fusion (F) gene- partial cds |
JX129806.1 |
NDV isolate NR_94 |
- |
Fusion (F) gene- partial cds |
JX129805.1 |
NDV/FALCON/RL/IS/2012/911 |
Falcon |
Fusion protein (F) gene- partial cds |
KF650624.1 |
NDV/CANARY/Je/IS/2012/875 |
Canary |
Fusion protein (F) gene- partial cds |
KF650623.1 |
NDV/CHICKEN/Hol/IS/2012/870 |
Chicken |
Fusion protein (F) gene- partial cds |
KF650622.1 |
NDV/PARROT/BS/IS/2012/84 |
Parrot |
Fusion protein (F) gene- partial cds |
KF650619.1 |
NDV-Ck-Pakistan-NARC-13N20-2013 |
Chicken |
Fusion protein (F) gene- partial cds |
KC811835.1 |
NDV-Ck-Pakistan-NARC-13N94-2013 |
Chicken |
Fusion protein (F) gene- partial cds |
KC811831.1 |
NDV-Ck-Pakistan-NARC-12N152-2012 |
Chicken |
Fusion protein (F) gene- partial cds |
KC811830.1 |
NDV-Ck-Pakistan-NARC-12N148-2012 |
Chicken |
Fusion protein (F) gene- partial cds |
KC811814.1 |
NDV-Ck-Pakistan-NARC-12N80-2012 |
Chicken |
Fusion protein (F) gene- partial cds |
KC811813.1 |
NDV-Ck-Pakistan-NARC-28991-2012 |
Chicken |
Fusion protein (F) gene- partial cds |
KC811812.1 |
NDV/ PARROT/BS/IS/2012/841 |
Parrot |
Fusion protein (F) gene- partial cds |
KF650619.1 |
NDV-Ck-Pakistan-NARC-13N20-2013 |
Chicken |
Fusion protein (F) gene- partial cds |
KC811835.1 |
NDV-Ck-Pakistan-NARC-13N94-2013 |
Chicken |
Fusion protein (F) gene- partial cds |
KC811831.1 |
NDV-Ck-Pakistan-NARC-12N152-2012 |
Chicken |
Fusion protein (F) gene- partial cds |
KC811830.1 |
NDV-Ck-Pakistan-NARC-12N148-2012 |
Chicken |
Fusion protein (F) gene- partial cds |
KC811814.1 |
NDV-Ck-Pakistan-NARC-12N80-2012 |
Chicken |
Fusion protein (F) gene- partial cds |
KC811813.1 |
NDV-Ck-Pakistan-NARC-28991-2012 |
Chicken |
Fusion protein (F) gene- partial cds |
KC811812.1 |
NDV-Ck-Pakistan-NARC-12N126-2012 |
Chicken |
Fusion protein (F) gene- partial cds |
KC811811.1 |
NDV-Ck-Pakistan-NARC-28789-2012 |
Chicken |
Fusion protein (F) gene- partial cds |
KC811810.1 |
NDV-Ck-Pakistan-NARC-28859-2011 |
Chicken |
Fusion protein (F) gene- partial cds |
KC811809.1 |
Table 2 Genbank accession data of the reference NDV strains aligned to the NDV field strains isolated from the four avid species (NCBI Blast-BLASTIN 2.2.29+)
Generally, it is acceptable that sequence of as little as 250bp always give meaningful convenient phylogenetic analysis compared to much longer sequences.18,19 Nonetheless, in this study, only 150bp of the totally amplified cDNA of the F-gene was successfully used to reveal results for molecular pathotyping and phylogenic analysis of the 216bp-amplified cDNA of the isolated NDV strains. The reverse is true for a convenient and precise genetic characterization studies.18,19 Previous investigations have established that, the fusion protein is likely one of the major proteins that play an important role indetermination of the genetic variations among different NDV strains, and that, it is more sensitive than the internal genes demonstrating genetic characteristics.20,21 For that, further future molecularly established investigations using F-gene primer sets of a convenient length is recommended to cast light on the biology, ecology, and epidemiology of the isolated NDV strains and the newly expected emerging strains as well.
The fact that falcons play a significant role in the traditions of Gulf citizens is very observable in Qatar State. Since, houbara, quails and pigeons are the common birds used as a main raw meat source for feeding falcons, and that they are highly ND-susceptible and reservoir birds, reared in mixed communal backyards and traditional farms, perpetuation of NDV in the country is anticipated. In addition, increased possibilities of emergence of new virulent virus mutants are also expected. Accordingly, much attention and more strict health measures are recommended to guard the growing investment in the country’s poultry industry. The cross-boundary nature of falcons as a hunting bird will also warrants additional health measures. Control measures based on identity of the nature of the candidate vaccine to combat the incriminated virus in the region should be escorted carefully.
This investigation was conducted in Virology and Molecular Biology Units, Veterinary Laboratory, Department of Animal Resources, and was finalized in the Department of Population Medicine, College of Veterinary Medicine, and University of Cornell, USA. The authors are grateful to the Director of the Department of Animal Resources and the Head of the Laboratory for permission to use those facilities. Thanks are extended to Professor Hussni OM for offering a venue to use Cornell facilities. Due thanks goes to the technical staff for helping during the study.
None.

©2014 Abdulla, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.