Journal of
eISSN: 2373-6453


Research Article Volume 8 Issue 2
1Kenya medical Research Institute (KEMRI), Kenya
2Jomo Kenyatta University of Agriculture and Technology (JKUAT), Kenya
Correspondence: Mainga O. Robert. Kenya Medical Research Institute (KEMRI), Centre for Virus Research, P.O. Box 54840-00200, Nairobi, Kenya, Tel +254-20-2722541, Fax 254-20- 2720030
Received: March 25, 2020 | Published: May 6, 2020
Citation: Robert MO, Eddy OO, Peter BK, et al. Characterization of Hepatitis C virus circulating among injecting drug users (IDU) in Kilificounty, Kenya. J Hum VirolRetrovirolog. 2020;8(2):23-30. DOI: 10.15406/jhvrv.2020.08.00217
Background: Hepatitis C virus is a major global health problem estimated to infect over 170 million people globally with the most common route ofinfection being injecting drug use (IDU). Treatment for HCV infection has traditionally been shown to be genotype specific; however the available drugs are still expensive and out of reach in many developing countries. To improve on monitoring, there is need to continuously document the genotypic burden and epidemiology in different populations arises.
Objectives: This study aimed to determine the circulating genotypes and link the data to the socio-demographics of injecting drug users in Kilifi County along the Kenyan coastline.
Methods: Using a random sampling method, this was a cross-sectional prospective study conducted among 127 injecting drug users, whereby ethical clearance was sought from Kenyatta National Hospital/ University of Nairobi Ethical and Research review committee (KNH/UON-ERC), and Reference number P366/07/2017 on 25th September 2017. Serology for HCV was done followed by nucleic acid amplification and eventual genotyping. Socio-demographic data was collected using questionnaire administered at the sites.
Results: A total of 28 (23 males and 5 females) samples out of 127 samples were positive for HCV giving a prevalence of 22.1%. The modal age group was 25- 49 years. Of the positive samples, 11 were amplified by PCR, all from the male IDUs. Prevalent genotypes included genotype 1a (13%) and genotype 4a (87%). Both genotype 1a and 4a were only found in males.
Conclusion: Although it is the first time to report HCV4a in Kilifi town and Mtwapa as compared to Watamu and Malindi which had both HCV1a and HCV4a. Tourists born in countries where HCV prevalence is high are supposed to be screened before accessing entry to Kilifi County. No clinical trial was conducted. The study reveals the burden of HCV infection among IDUs inKilifi County. The Government should formulate policies for intervention on testing and treatment of HCV in Kilifi County targeting IDUs in order to minimize spread to other populations.
Keywords: characterization, hepatitis cvirus, genotyping, prevalence
Bp, base pair; CI, confidence interval; CSW, commercial sex workers; dNTPS, nucleoside triphosphate; EDTA, ethylenediaminetetraacetic acid; DNA, deoxyribonucleic acid; HCC, hepatocellular cancer; HCV, Hepatitis C virus; HCV1a, Hepatitis C virus Genotype one Subtype a; Hcv4a, HCVFour Subtype a; IDUs, injecting drug users; KANCO, Kenya AIDS NGOs Consortium; KEMRI, kenya medical research; MEWA, muslim education welfare association; n, participants who are either positive or negative; n/N, percentage of participants; N, number of participants; OR, odds ratio, P, P value; PCR, polymerase chain reaction; RNA, ribonucleic acid; RT-PCR, ribonucleic acid; SPSS, statistical package for the social sciences; TE buffer, buffer used to solubilize DNA or RNA, while protecting it from degradation; µl, microliters; 5’UTR, five prime untranslated region; WHO , world health organization; ZR, zymo research
Hepatitis C virus causes of liver cancer such as hepatocellular carcinoma (HCC) and lymphomas in humans.1,2 Hepatitis C is a global health problem, over130 million people are chronically infected yearly.3 Studies show that every year, approximately 308,000 deaths occur due to liver cancer while up to 758,000 deaths are estimated to occur due to liver cirrhosis.4Approximately 80% of HCV infected persons will develop chronic hepatitis. Eleven percent will progress to liver cirrhosis for the period of 20-year time interval. Life-threatening will be as a result of liver failure which leads to hepatocellular carcinoma.5,6Blood products, needle or syringe sharing among members of intravenous drug parties or undergoing a needle stick by health workers leads to transmission of HCV. Other risk factors are high-risk sexual behaviors, tattooing, reused and unsterilized dental and surgical instruments, and unsterilized laboratory equipment.7About 80% of HCV infected persons are asymptomatic. Infected persons with acute HCV exhibit symptom ranging from fever, joint pain and jaundice. Asymptomatic individuals are more difficult to identify.8
Most IDUs with persistent infection are unaware of the infection, screening programs to identify patients will be required to prevent silent progression of the disease.9–11In most case HCV is often first diagnosed in late stage. Due to slow and silent onset, many patients are unaware of their infection and at least 40% cases remain undetected. Chronic hepatitis C infection is difficult to assess, because it is frequently subclinical.5 Patients with chronic hepatitis C are at risk of cirrhosis and hepatocellular carcinoma and their contacts (especially injecting drug users, IDUs and commercial sex workers CSWs) at risk of acquiring the infection through exposure to the virus.12Given this situation, it appears that there is no clear understanding of the contextual factors that continue to fuel the upsurge in HCV infections among the key populations such as IDUs and CSWs.13Molecular genotyping of HCV identified in patients is necessary, for proposing therapeutic options.6 It is assumed that some genotypes are more common in certain areas or groups of people. The high number of chronically infected individuals, the burden of disease and the absence of a vaccine indicates that treatment will form part of the control of the disease.14 The aim of this study was to determine the prevalence, social characteristics and HCV genotypes circulating among IDUs in rehabilitation centers in Kilifi County Kenya.
Study site and study design
This study was conducted in Kilifi County located within the Kenyan coastline. The design was cross sectional. Participants were recruited from harm reduction/rehabilitation centers within Malindi, Watamu, Kilfi town and Mtwapa {note that Kilifi town and Mtwapa is one centresplited into two called Muslim Welfare Association (MEWA)} located at Kilifi County.
Sample collection, transportation and storage
After informed consent, Socio-demographic data was captured using questionnaire which was administered at the facilities. The data captured included, name of centers, date of birth, residential area, marital status, religion, occupation and the level of education. Further five milliliters of venous blood was collected into EDTA vacutainer tubes. They were then transported in dry ice from rehabilitation Centers to KEMRI laboratories for analysis.15
Serological tests
Viral RNA was extracted from 170 µl of serum byusing Zymo Research Kit (IrvineCA, U.S.A.) according to the manufacturer’s instructions. The RNA pellets were resuspended in 60 µl of TE buffer. The RNA concentrations were measured in ng/µl using NanoDrop spectrophotometer (ThermoFisher Scientific, Wilmington, Delaware, USA).
Polymerase Chain Reaction (RT-PCR) and Sequencing reaction
Nested RT-PCR was carried out using Zymo Research PCR Kit (IrvineCA U.S.A.)and GeneAmp PCR system 9700.18 In reverse transcription and PCR reactions, both 5′UTR universal primers: forward KY80 (5’-GCA GAA AGC GTC TAG CCA TGG CGT-3’) and reverse KY78 (5’-CTC GCA AGC ACC CTA TCA GGC AGT-3’) were used. 35 cycles of PCR (94°C for 30 seconds, 54°C for 1 minute, and 68°C for 2 minutes) were performed HCV PCR products of 244 bp were visualized, and electrophoresed on 1.5% agarose gel.9,19 The positive amplicons were purified using the ZR DNA sequencing clean up kit.20 (Zymo, Irvine, CA, USA) according to the manufacturer’s protocol to eliminate unincorporated primers and dNTPs.21 The purified amplicons were sequenced with Big Dye kit, and the ABI PRIS M 3100 genetic analyzer (Applied Biosystems) according to manufacturer’s instructions.
Sequence analysis
Samples were aligned with the representative sequences for each major genotype and subtype selected from the HCV database and Gene Bank using the Multiple Sequence Alignment Program, ClustalW. Homology and evolutionary distance pair-wise comparisons for percent nucleotide were made.21The phylogenetic analysis of HCV isolates was performed with MEGA7.0.14software.
Statistical Methods
Statistical Package for the Social Sciences (SPSS) version 20 software (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. Non-parametric Mann-Whitney U test was used to compare between genotypes with respect to quantitative variables. Chi square was used to analyze association between social demographics and circulating HCV genotypes. A p-value of less or equal to 0.05 was considered statistically significant (Valentine et al., 2018).
Demographic characteristics of study participants
A total of 127 IDUsregistered patients were recruited for the study {107 (84.2%) were male and 20 (15.7%) were female}. The samples in each site were as follows; Malindi, (n =35), Watamu (n =43), and Kilifi and Mtwapa (n =49) respectively. The mean age for the three centers was found to be 33.7 years.
Prevalence of HCV among study participants
Anti-HCV positive included 10 IDUs from Kilifi, 4 from Watamu and 14 from Malindi making a total of 28 out of 127 participants. Twenty three IDUs (82.1%) were male while 5 (17.9%) were women. Of the 28 IDUs, 11 (8.7%) male were confirmed to be HCV positive using PCR as compared to none for female IDUs.A total 28 participants screened, seroprevalence was established at 22.1%. With the highest prevalence found in males (18.1%; n=23) and low in females at (3.9%; n=5) (P=0.004). The seroprevalence in Malindi was 40%, Watamu 9.3% and Kilifi and Mtwapa 20.4% respectively(Table 1).
Rehabilitation centers |
Overall |
P Value |
|||||
KILIFI, n=49 |
WATAMU, n=43 |
MALINDI, n=35 |
|||||
Variable |
n (%) |
n (%) |
n (%) |
n (%) |
|||
Rapid test |
|||||||
Negative |
39 (79.6) |
39 (90.7) |
21 (60.0) |
99 (78.0) |
0.004 |
||
Positive |
10 (20.4) |
4 (9.3) |
14 (40.0) |
28 (22.1) |
|||
PCR |
|||||||
HCV1a |
0 |
1 (33.3) |
2 (66.7) |
3 (27.3) |
0.121 |
||
HCV4a |
5 (100.0) |
2 (66.7) |
1 (33.3) |
8 (72.7) |
|||
Table 1 The table above reveals the Prevalence of Hepatitis C Virus (HCV) which is 22.1% and its Genotypes among study participants in Kilifi County
Serology outcome and demographic characteristics
The highest number of injecting drug users was in the age group of 31-35(39%),it also had a higher number of IDUs who tested positive 8 (20.5%) as compared to other age brackets. In the study, 35 IDUs sampled were married, 68 single and 24 divorced. (OR=1.0, 3.0, 2.0. P = 0.065, 0.330 and CI: 0.9-9.7, 0.5-8.6 respectively). Those of the Muslim faith had higher number of HCV infections, 16 compared to Christians, 12 and pagans zero. (OR= 1.0, 1.4 and P= 0.497, 0.309 respectively). In education, Lower primary had 13, Upper primary 6 and Secondary and Tertiary 9 (OR=1.0, 0.8, 2.1, P = 0.682, 0.146. CI: 0.3-2.3) for upper primary, CI: 0.8-5.8 for secondary and tertiary respectively).Tout/drivers/riders group of IDUs had highest HCV infection 12, (OR=1), Beach boys were5, Fishermen were 3 and other IDUs with different occupations were 7 respectively. (OR=1.0, 1.3, 0.8, 0.9. P= 0.688, 0.712, 0.831 CI: 0.4-4.3, 0.2-3.1, 0.3-2.6 respectively)(Table 2).
Variable |
N |
(n/N) % |
Positive |
Negative |
OR (95%CI) |
P value |
Age |
3 |
66.7 |
2 (66.7) |
1 (33..3) |
1.0 |
0.171 |
Gender |
107 |
21.5 |
23 (21.5) |
84 (78.5) |
1.0 |
0.729 |
Marital status |
35 |
11.4 |
4 (11.4) |
31 (88.6) |
1.0 |
0.065 |
Religion |
77 |
0 |
16 (20.8) |
61 (79.2) |
1.0 |
0.497 |
Education |
65 |
20.0 |
13 (20.0) |
52 (52.5) |
1.0 |
0.682 |
Occupation |
55 |
21.8 |
12 (21.8) |
43 (78.2) |
1.0 |
0.688 |
Table 2 The table above shows correlation between Serological outcome and demographic characteristics of Participants in Kilifi County
Key: N, number of participants, n, participants who are either Positive or negative (n/N) percentage of participants, OR, odds ratio, CI, confidence intervals
Polymerase chain reaction (PCR) and genotyping
The 28 samples which were positive were subjected to PCR, 11 (39.3%) were confirmed positive (all males) and none of the 5 female IDUs was positive. The 11 samples were then subjected for sequencing and results gave 3 out of the 11 samples as HCV Genotype 1 subtype a (HCV1a) while 8 were HCV Genotype 4 subtype a (HCV4a)(Table 3).
Variable |
HCV1a (%) |
HCV4a (%) |
P value |
Age |
|
0 |
|
Gender |
3 (27.3) |
8 (72.7) |
|
Marital status |
2 (40.0) |
3 (60.0) |
0.452 |
Religion |
0 |
0 |
|
Education |
1 (25.0) |
3 (75.0) |
0.145 |
Occupation |
2 (40.0) |
3 (60.0) |
0.488 |
Table 3 The table above shows correlation of Polymerase Chain Reaction (PCR) and Genotypes with participant demographic characteristics in Kilifi County. It reveals the percentages of Hepatitis C virus Genotype one Subtype a {HCV1a (%)} and Hepatitis C virus Genotype four Subtype a {HCV4a (%)} among the IDUs in Kilifi County
Phylogenetic tree construction
With reference of Full-genome consensus sequences for different HCV genotypes (including genotypes 1, 2, 3, 4, 5 and 6), there was only genotype 1 and 4 (Figure 1).The rest there was no evident on the tree because there were no associated isolates. Maximum composite likelihood algorithms were utilized, and phylogenetic trees were constructed by the neighbor-joining method. The reliability of different phylogenetic groupings was evaluated by using the bootstrap re-sampling test from the MEGA program (1,000 bootstrap replications).19,22–25
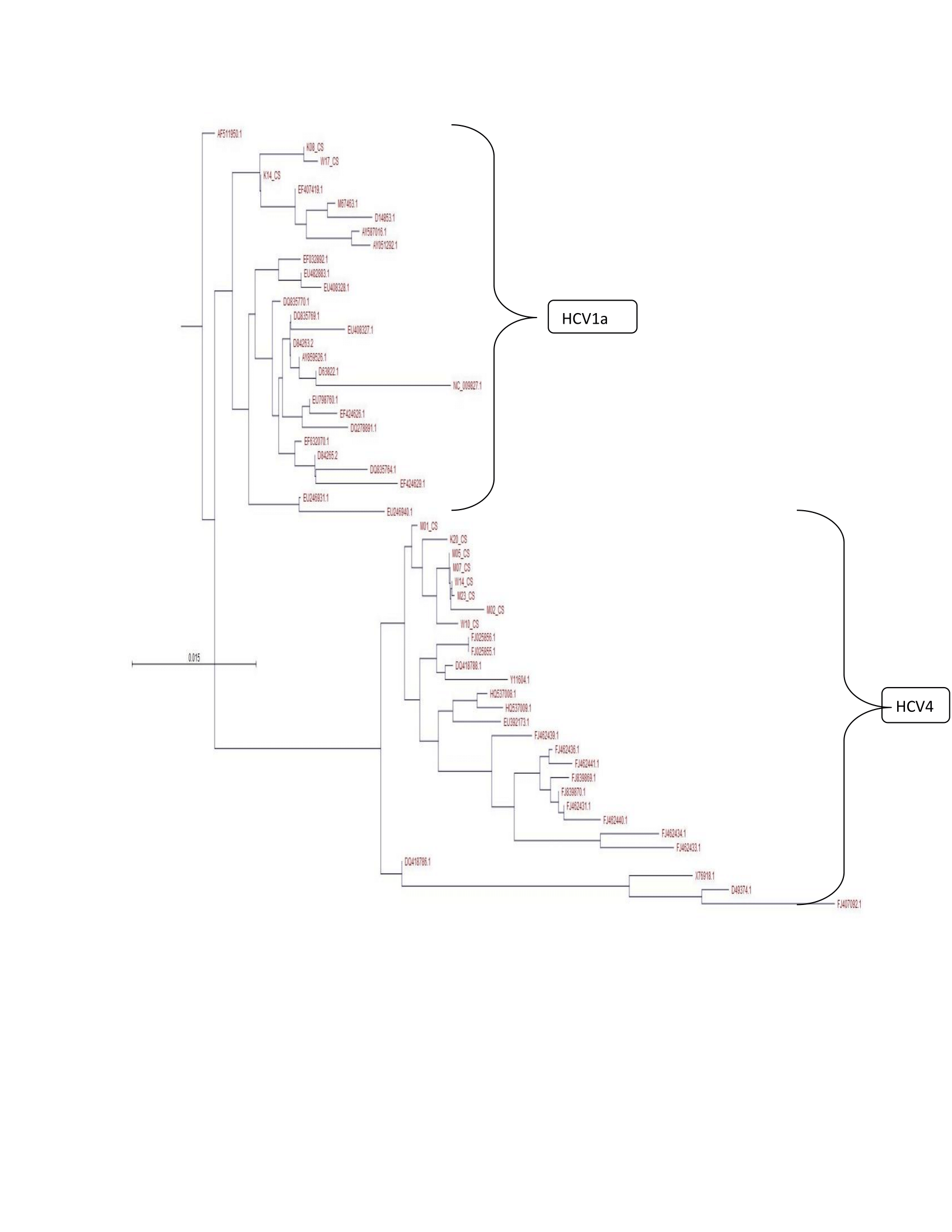
Figure 1 Phylogenetic neighbor-joining trees of Hepatitis C Virus (HCV) 5′UTR partial sequences in Kilifi County, it revealed two monophyletic clusters (bootstrap value,∼87) containing Hepatitis C Virus Genotype one Subtype a (HCV1a) and Hepatitis C Virus Genotype four Subtype a (HCV4a) among infected patients in the County.
In this study we have managed to establish a higher prevalence of HCV in male injectors107 (84.2%) than females 20 (15.7%). We have also shown a changing trend of HCV genotype distribution with genotype 4a becoming more prevalent in this region as opposed to the traditional genotypes 1a, genotype 1a can be seen in Watamu and Malindi but not in Kilifi, it also shows that it is the first time to report HCV4a in Kilifi. Hepatitis C patients were found to be illiterate, it shows that lack of knowledge about the disease and its precautions become a strong cause for disease. In this study, there is an increase of HCV infection as compared to the study which was done by Sacks-Davis R17in the entire Coast which showed a prevalence of 16.7%.We have shown that there is increased diversity along the coastal strip centers like Mtwapa which is part of Kilifi town both are under Muslim welfare association (MEWA).
Genotyping of hepatitis C virus (HCV) is considered an important tool for epidemiological and clinical studies and valuable marker for disease progression and response to antiviral therapy.26However, development of antiviral drugs, vaccines, and genotyping assays has a major impact on HCV this is because HCV has high levels of genetic diversity.19,27,28In this study, we have confirmed that only two genotypes circulating in Kilifi County; this can form a basis for more targeted control for the 2 subtypes in the region.Hepatitis C Virus diversity is very essential in response to antiviral therapy.21,29 More severe liver disease and more elevated liver enzymes have been highly associated with HCV genotype 1.30 The endemicity of HCV genotypes in some areas is revered with multiplicity and diversity of subtypes and strains. A good example is origin of genotypes 1 and 2 to West Africa and genotype 4 to Central Africa.31 This can be done by using a phylogenetic analysis to revealed two monophyletic clusters (bootstrap value,∼87) containing HCV infected patients in Kilifi County from which a partial 5′UTR sequence was available.
The largest cluster contained 87% of HCV sequences were identified as genotype 4, with isolates M01, M02, M05, M07, M23, K20, W10, and W14. This isolates clusters with isolates from: Portugal, France, Egypt, Cyprus, Southeast Asia, Middle East and Saudi Arabia. On the other hand genotype1a Isolates were K08, K14 and W17 Clusters with isolates from: America, Japan, Indonesia, China and India. Genotype diversity is particularly high in China and many Southeast Asian countries and also in Western Europe and Australia, perhaps as a result of population immigration from Africa and/or Asia.32In countries like these, vaccine efficacy at a population level would be dependent on the generation of cross reactive immunity; an alternative approach could also be the development of vaccines hosting different immunogens. In this study, HCV genotype 4a and genotype1a is predominant among the population of drug users, this suggest that there is a need for continuous monitoring of these for better treatment outcomes that entirely depend on HCV genotyping results. With global travel, increasing of tourist to Kilifi Count and the geographic zone of this region dictates the distribution of these genotypes in this region.33
Due to lack of vaccine and effective therapy, the prevention of HCV infection has been a great challenge to developing country and owns one-fifths of the world’s population.34,35World Health Organization Assembly approved on 2016 the Global Health Sector Strategy to eliminate hepatitis infection by 2030.36It introduced global targets for the care and management of HCV including 90% reduction in new cases of chronic hepatitis C, 65% reduction in hepatitis C deaths, and treatment of 80% of eligible people with chronic hepatitis C infections.37To achieve these goals, the country need to develop national policies based on reliable epidemiological evidence.38 In Kilifi County, Screening populations at risk and Counseling program to IDUs with history of sexual contact or sharing of items should be done to stop spread of HCV in the region. Tourists born in countries where HCV prevalence is high are supposed to be screened before accessing entry to Kilifi County.However, data are often outdated and conflicting, making evidence-based policy and resource allocation difficult.
This study reveals the burden of HCV infection among IDUs population inKilifi County. HCV infection seroprevalence rate seem to be increasing up to (22.1%) as compared to the previous studies 16.4% by Mwatelah17 and 22% by Akiyama,39this might become a major health predicament in future. Although screening and treatment is expensive, the Government should put control measures like conducting constant surveillance, educating, screening and treatment of the affected population in order to control the disease from the area.
I wish to express my sincere appreciation to my supervisors Dr. Eddy Odari, Dr. Raphael Lihana and Dr. Peter Borus for their guidance in the design and implementation of the study, overall supervision of the project. I would also like to thank Dr Charles Mwandawiro for providing funds for my laboratory work. I would acknowledge Jocelyn katunge (Chief youth development officer Kilifi County), FatmaJeneby (MEWA programme coordinator), Hussein Taib (MEWA programme supervisor), Alfred Jumaa (MEWA Clinical officer), BilhaMaina (KANCO Project coordinator), BeryneAgutu (Clinical Officer at Omari rehabilitation centre) and all the IDUs who made this project successful. Lastly, I thank Dr Allan Ole Kwara, BenardOsero, Vincent Rutto, James Gikunda and Robert Oira for their assistance.
Ethical consideration
Ethical clearance was sought from Kenyatta National Hospital/ University of Nairobi Ethical and Research review committee (KNH/UON-ERC), Reference number P366/07/2017 on 25th September 2017.
Consent
Consent to participate is attached together with the worksheet and Manuscript.
Availability of data and materials
It is available in IDUs worksheet that I have attached.
Conflicts of interest
There were no Conflicts ofinterest in this project.
Funding
The project was funded by Children investment fund foundation (CIFF). File Number: E- 2008. The owner of the project frontier to buy for me reagents since I was a student, I dint have a source of funds for my project.
Author’s Contributions
O. R. He is the author, his work was to collect samples, do laboratory work (both serological and molecular work) and writing of manuscript.
O. E. He supervised laboratory work, assisted in field work, review manuscript writing and data analysis.
K.P. His work was to review manuscript and administrative work.
K. A. His work was to review manuscript and laboratory work analysis.
G. J. His work was field work and laboratory analysis
K. V.His work was to assist in laboratory analysis.
S. C. He assisted in funding of the project, field work and administrative work.
W. R. He supervised laboratory work, review manuscript writing and data analysis.
All authors have read and approved this manuscript for publication.

©2020 Robert, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.