Journal of
eISSN: 2373-6453


Short Communication Volume 6 Issue 3
Department of Microbiology, University of Lagos, Nigeria
Correspondence: Adeyemi Adeleye, Department of Microbiology, Faculty of Science, University of Lagos, Akoka, Nigeria,
Received: March 01, 2017 | Published: December 10, 2018
Citation: Adeleye A. A review on torque teno virus. J Hum Virol Retrovirology. 2018;6(3):96-98. DOI: 10.15406/jhvrv.2018.06.00202
Torque teno virus (TTV) formally known as Transfusion transmitted virus is a single stranded DNA virus discovered in 1997 by Nishizawa and co-workers while attempting to shed light on the possible causes of hepatitis of non A-G etiology in some Japanese patients.1 The patients presented with post transfusion hepatitis of unknown etiology. When investigated, the serum of these patients was found to contain a DNA sequence which was not similar to any sequence in the Gene bank. This new DNA sequence was then identified as belonging to a novel viral agent which was suspected could be responsible for the development of hepatitis symptoms in this patient.1 Further investigation also revealed that the patients had elevated alanine aminotransferase levels (ALT) after transfusion.
Genomic organization of TTV
TT virus genomic size varies among isolates. Approximately the genome is about 3.8kb.2 with a diameter of around 30-50nm. The genome has a coding region of approximately 2.6kb and an untranslated region (UTR) of 1.2kb. It contains a GC-rich region of about 120 nucleotides which is present in all isolates.2 The genome has three large open reading frame (ORF) which encodes proteins with 770, 202 and 105 amino acids.3 The ORFs encoding TT virus proteins are located in the complementary DNA strand, the plus strand. Despite TT virus sequence variability among isolates, the ORF1 is found in all isolates and encodes a capsid.4 The ORF1 sequence varies at the amino acid level among TT virus isolates with an amino acid differences of about 50-70% in these isolates.5 These are not uniformly distributed since ORF1 contains hypervariable regions (HVRs) where mutation occurs (Figure 1).6 It was believed that TT virus could escape immunological responses by mutating this HVRs.7
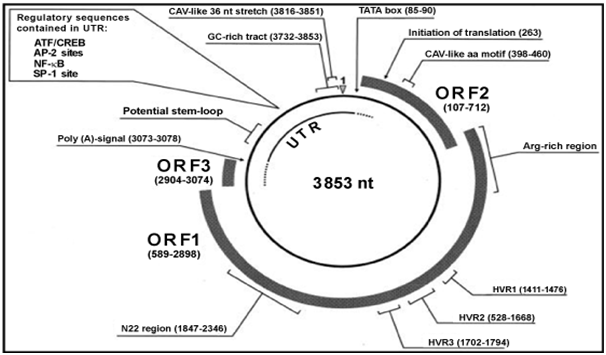
Figure 1 N=57; Epidemiological distribution of the pathological fractures, traumatic fractures, and nonunion.
TT virus replication, genetic diversity and transmission
The mechanism by which TT virus replicates is still unknown. It is assumed that replication in TT virus is similar to that of other circular single stranded DNA viruses.7 It is yet unknown what cells support replication of TT virus. Presence of TT virus mRNA form and dsDNA has been detected in various human organs and tissues.8 This indicates that they could be active sites for replication of TT virus.8 The presence of TT virus infection has been based on the detection of its DNA in serum. TTV is known to have a high genetic variability with at least 40 genotypes,9 exhibiting greater than 30% nucleotide differences from one another, and five major phylogenetic groups showing greater than 50% divergence nucleotide sequences.3 The genome shows greater variation at ORF level than in the UTRs and also have a high rate of mutation which was never observed in a human virus with such a small genome size. It was suggested that only a minor fraction of the TT viruses have all the components required for a successful infection or that a co infection of isolates brings the necessary component together.10 TTV infection is known to cut across races, gender, age or class. It is globally dispersed and can be detected in blood and blood products, in semen, water, cord blood, faeces, breast milk, throat swabs, nose swabs, saliva etc. suggesting that it can be contracted from environmental sources. TTV infections are extremely prevalent both in healthy individuals and diseased patients but no definitive causal association with TTV infection has been found for any of the diseases investigated. The ubiquitous nature of this virus raises the speculation whether the virus is pathogenic, opportunistic, a cofactor of other infections or a modulator of immunity that can promote other virus to be infectious.11 It is also believed that it is possible that some genotypes of TTV are pathogenic. Ever since the discovery of this virus, several studies have being done to understand the biology of this virus, its mechanism of infection and replication, association with any disease, the immunology of infection and the prevalence in different human tissues and study groups. The true identity of TT virus will still remain a puzzle until a suitable cell culture system is developed to study its biological characteristics.
Other variants of torque teno virus
In 2000, torque teno mini virus (TTMV) was accidentally discovered in serum of blood donor by PCR using TTV- specific primers that partially matched homologous sequences in TTMV but generated a noticeable shorter amplicon than that expected for TTV.12 Although TTV and TTMV have a common presumed genomic organization, they exhibit high dissimilarity in terms of genomic length and genetic identity. TTMV was found to be about 2.8-2.9kb in size. A third group in the genus Anellovirus was also discovered having a genomic organization resembling those of TTV and TTMV with a circular DNA genome of 3.2kb and was provisionally designated as torque teno midi virus (TTMDV).
Prevalence of TTV in different groups
A large number of epidemiological studies clearly pointed out the global distribution of the virus (Africa, North and South America, Asia, Europe, Oceania) in rural and urban population with more than 80% of the population being affected in some areas.11 TTV DNA detection rates are highly dependent the very high genetic variability of TT virus, the choice of primer used, the target gene, geographical region, standard of living.13 For example, prevalence in healthy rural populations in Africa was reported in Tanzania, 74%; Gambia, 83% and Ghana, 88%.14 In Saudi Arabia, epidemiological studies and genotyping of TT virus isolated from blood donors and hepatitis patients using ORF1 primers and 5’ UTR primers revealed that healthy blood donors had TT virus DNA of (5.5% and 50.5%) and overall (4.8% and 56.4%) among the hepatitis patients respectively. In the UAE the result obtained showed that TT virus infection in healthy nationals and those with HBsAg or antibody to HCV were 34.9% of 106 patients, 97.9% of 48 patients and 95.7% of 46 patients respectively, compared to 89.1%(115/129), 89.2%(66/74) and 84.8%(39/46) respectively in non-nationals.15 In Nigeria, in an ongoing study by Elesinnla et al, the prevalence of TTV among 130 blood donors and 130 HIV patients was put at 26% and 65% respectively. Likewise 93% and 88% of the HIV positive patients were positive for TTMV and TTMDV while 95% and 94% of the donors were positive for TTMV and TTMDV respectively.16 In a similar study in USA, it was discovered that the prevalence of TTV and TTMV infections in the HIV/HCV group and the HIV group were significantly higher than the control group (p<0.05). Furthermore, it was stated that the TTV and TTMV co infections were found in 92.2% in the HIV/HCV group 84.2% in the HIV group and 63% in the control group.17
Torque teno virus (TTV) formally known as Transfusion transmitted virus is a single stranded DNA virus discovered in 1997 by Nishizawa and co-workers while attempting to shed light on the possible causes of hepatitis of non A-G etiology in some Japanese patients. The patients presented with post transfusion hepatitis of unknown etiology. Further investigation also revealed that the patients had elevated alanine aminotransferase levels (ALT) after transfusion.
None.
The author declares there is no conflicts of interest.

©2018 Adeleye. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.