Journal of
eISSN: 2373-6453


Ebola virus diseases (EVD) have been a global challenge for almost 40 years to scientists around the globe. It is a violent pathogen causing outbreaks in various parts of the world. Lately in 2014 the outbreak began in the republic of Guinea and spread to the republic of Liberia and the Sierra Leone. The 2014 West Africa EBOLA outbreak remains to be most dreadful in medical history with both the numbers and fatalities. Due to meager progression of efficient cultivation of EBOLA virus, studies are limited and a wealth of information is not available. There is lack of approved drugs and vaccines and studies are limited to cell line and animal models. Biosafety concerns associated with containment are quite strong, posing limitations for studying Pathophysiology and virulence determinants. Despite limitations, incubation period, virus life cycle and a few molecular players involved in replication as well as the cytoskeleton are known. Prophylaxis measures are clearly indicated and practiced presently. Most of the research outcome has focused on preventing entry of virus either by the use of peptide base inhibitors, and monoclonal antibodies. Antivirals that target RNA synthesis of EBOLA virus, such as; small molecules and oligonucleotide derivatives are being developed. Alternative approaches of modifying symptoms rather than modifying the EBOLA virus are also being considered. Prevention of RNA synthesis and blocking viral entry approaches are promising and summarized in this review.
Keywords: EBOLA virus, EVD, Biosafety measures, Antivirals, MABs
NTD, Neglected Tropical Disease; EBOV, EBOLA Virus; MARV, Marburg Virus; SEBOV, Sudan Ebolavirus; BDBV, Bundibugyo Ebola Virus; REBOV, Reston Ebolavirus; ZEBOV, Zaire Ebolavirus; EVD, Ebola Virus Disease; TAFV, Tai Forest Ebola Virus; MLV, Marine Leukaemia Virus; VSV, Vesicular Stomatitis Virus
EBOLA virus disease (EVD), a neglected tropical disease (NTD), has always been a challenge and a global menace since its discovery in 1976.1 EBOLA virus (EBOV) and Marburg virus (MARV), members of the Filoviridae family, are known as emerging and re-emerging zoonotic pathogens causing acute hemorrhagic fever with a high case-fatality rate in humans (up to 90%). EBOLA virus is a virulent, lipid-enveloped, negative stranded RNA virus that belongs to the viral family Filoviridae. Five members of the Filoviridae family: Zaire ebolavirus(ZEBOV), Sudan ebolavirus(SEBOV), Côte d’Ivoire ebolavirus, Bundibugyo ebola virus (BDBV) and Reston ebolavirus(REBOV). In February of 2014, the outbreak began in the republic of Guinea and the virus spread to the republic of Liberia and the Sierra Leone. The 2014 West Africa EBOLA outbreak is believed to be the most terrible in medical history with regards to both the number of human cases and fatalities.2-9 Presently, Vaccines against Ebola virus disease are an urgent international priority.10 However, at present; no licensed vaccines are available, despite promising results for several candidate vaccines in non-human primate studies and phase 1 trial.2-16 EBOLA investigators have developed several research models in both cell culture17-20 and animals.21-24 EBOVs can be cultured using the Vero E6 cell line, and this model provides the entire virus replication cycle for drug research.17 Recently, a recombinant, replication-competent vesicular stomatitis virus-based vaccine expressing a surface glycoprotein of Zaire Ebola virus (rVSV-ZEBOV) is a promising Ebola vaccine candidate reported by Henao-Restrepo AM et al.25
The prevention of EVD requires more awareness of the pathology of the ailment, especially the role of wildlife, especially bats, in the spread of EBOLA virus to humans. The present review is an attempt to summarize various essential aspects of EVD.26
The virus
The EVD, (Former-EHF) is a severe condition caused by a virus belonging to genus Ebola virus, family Filoviridaeand order Mononegavirales. The family Filoviridaeis comprised of one genus, Filovirus, which contains two species, morphologically identical but serologically distinct: Marburg virus and Ebola virus. There are five EBOLA subtypes BDBV, ZEBOV, REBOV, SEBOV and Tai Forest ebola virus(TAFV) which vary in pathogenicity, antigenicity and genomic constitution.27 The identification and successful isolation of Marburg virus from the cave-dwelling fruit bat Rousettus aegyptiacus.28 Ebola virus might endure as an asymptomatic or subclinical infection in the reservoir species, with little or no transmission, and might be intermittently provoked through a suitable stimulus. The stimulus might be stress, co-infection, change in food sources and pregnancy as displayed in vivoand in vitroinvestigations.29,30 Mammalian species, including NHP, vulnerable to infection are considered as the dead end hosts.
Structure of EBOLA virus
EBOLA virus is an infectious, intracellular parasite. EBOVs have a threadlike shape, with a uniform diameter of 80 nm (Figure 1).31 The typical length of a virion with peak infectivity is about 1200 nm.32 The virus genome consists of a single 19 kb strand of negative sense RNA with seven viral genes that are transcribed by the viral RNA dependent RNA polymerase present in the virion. The single strand of RNA is covered by the helically arranged viral nucleoproteins NP and VP30, which are linked by matrix proteins VP24 and VP4 to the lipid bilayer that coats the virion.33 The EBOLA virion genome enclosed in bacilliform particles, encodes seven genes: nucleoprotein, VP35, VP40, GP, VP30, VP24, and RNA-dependent RNA polymerase (L).The major nucleoprotein (NP) and virion protein 30 (VP30, minor nucleoprotein),a viral transcription activator, are required for RNA encapsulation (Figure 1).34,35
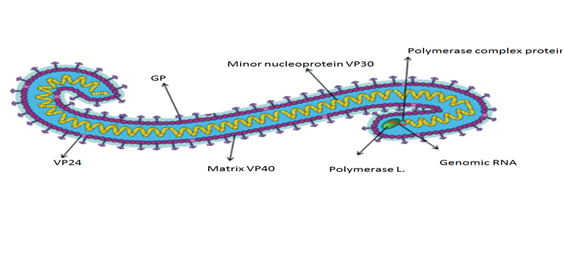
Figure 1 Ebola virus structure.93
Where - VP: virion protein; GP: Glycoprotein.
The virion protein 35 (VP35) links NP with the viral RNA dependent RNA polymerase (RdRP, polymerase L) to form a complex for replication and transcription.36,37 EBOLA virus goes through 6 phases of viral replication: attachment, penetration, uncoating, replication and expression, maturation, and release/delivery of virus. The matrix proteins, virion protein 40 (VP40) and virion protein 24 (VP24) are associated with virus assembly and budding, and also have specific affinity for membranes.38 Glycoprotein (GP) spikes, which are embedded on the virion surface, mediate virus entry39 (Figure 2). The GP gene also encodes soluble GP (sGP) and small soluble GP (ssGP), which are secreted from the host cell.1 On the virion surface glycoprotein (GP), member of class I viral fusion proteins is synthesized as a single polypeptide precursor called Pre-GP. Pre-Gp then undergoes modifications by N-glycosylation and O-glycosylation into a fully glycosylated form, GP0. The GP0 is cleaved in the later-Golgi by furin-like proteases into GP1 and GP2. On the virion surface GP is present as a homotrimer and contains essentially two subunits: a large surface subunit (GP1) that binds to the cell surface receptor and a transmembrane subunit (GP2) with transmembrane domain, which mediates the virus-host membrane fusion. Importance of GP2 transmembrane domain in enhancing membrane permeability is essentially important in pathogenesis.
GP1 is responsible for binding with a variety of host cell surface factors, and covering the receptor binding domain (RBD) under them.40 Three fusion peptides on the N terminal of the GP2 insert into the endosomal membrane, launching the six-helix bundle (6-HB) formation between the N- and C-terminal heptad repeats (NHR and CHR, respectively) and viral-host cell membrane fusion, in a manner similar to that mediated by other type I viral membrane proteins.41,42 Sequentially, the genome and RNA synthesis machinery is released into the cytoplasm for another cycle of transcription, protein translation, genome replication, and virion assembly.1 EBOVs prefer to use mononuclear cells in the early stage of infection, such as tissue macrophages, monocyte and dendritic cells, for rapid virus replication.1,43 EBOLA viruses have a tropism for a large range of cell types like hepatocytes, kidney cells and other epithelial cells. The replication is mainly because of the viral proteins that antagonize the host interferon response.
Cytokines released from infected cells recruit more mononuclear cells to the initial infection site, in turn amplifying infection and apoptosis of mononuclear cells. At the same time, virions are systemically spread through blood circulation. The glycoprotein and RNA synthesis machinery, which play important roles in viral entry and RNA replication, respectively, are promising drug targets for EBOLA therapies.
EBOLA virus culture:
Studies analyzing EBOLA virus replication have been severely hampered by the extreme pathogenicity of the virus. Numerous factors such as cell substrate, virus strain, expression system, medium, cultivation system, cultivation method, and scale need consideration for the growth of the EBOLA virus. To permit analysis of the host range and function of the EBOLA virus, various systems were developed. In 1992, Titenko et al.44 studied the EBOLA-Zaire virus production in Vero and BGM cells. The cell monolayer destruction of 80-90% was seen at a low multiplicity of infection in 7-8 days after virus inoculation. At a multiplicity of infection of 0.01 PFU/cell, the maximum virus titter of 10 PFU/ml was found 7 days post infection.44 Similarly Oestereich L (2014) observed T-705 suppressed replication of Zaire EBOV in cell culture by 4 logs 10 with an IC90 of 110μM. Mice lacking the type I interferon receptor (IFNAR (-) (/) (-)) were also used as in vivo model for Zaire EBOV-induced disease.26 In addition, a system of pseudo typing the glycoproteins of EBOLA into marine leukaemia virus (MLV) was developed.45
The life cycle and pathophysiology
In the first EBOLA out breaking the Democratic Republic of the Congo, in Yambuku, the reuse of unsterilized needles and syringes was a causative factor in the transmission of the disease. In the Kikwit outbreak, several clinic employees became infected because of improper barrier measures.46,47 The preliminary replication happens in monocyte, macrophages and dendritic cells and via these cells the virus is distributed to lymph nodes, spleen, liver and other organs. Increases in interleukin-2, interleukin-10, tumour necrosis factor, interferon-alpha and gamma were observed in fatal EVD cases. The virus is capable of persisting on objects for several hours in a dried state and can persist for a few days within body fluids.48,49 More noteworthy effects are micro vascular damage, changes in vascular permeability and activation of the clotting cascade. The impairment of platelets and endothelial cells causes disruption of fluid balance and homeostasis. In addition, the virus is considered to hide immunological function.50 The virus has been found in semen for up to 7 weeks after recovery from the illness, indicating the probability of a sexual mode of transmission. EBOLA virus infection may also spread through breast milk of women after recovery and it is unknown when it is prudent to breast feed again. Otherwise, people who have recovered are not infectious.51 Aerosol transmission has been observed among monkeys infected with the Reston and Zaire subtypes of EBOLA virus. EBOLA virus has also been identified in alveoli of experimentally infected monkeys.52 The spread array during an epidemic in humans does not advocate the respiratory route of transmission. No evidence suggests the role of insects in transmission of EVD.52,53 Dead bodies remain infectious; thus, people handling human remains in procedures such as traditional burial rituals or more modern processes such as embalming are at threat.54
The disease has an incubation period of 2-21 days(average 4-10 days) followed by symptoms including fever followed by headache, fatigue, dysphasia or odynophagia, abdominal pain, myalgia, sore throat, cough, anorexia, nausea, vomiting and diarrhea.50 A conjunctiva infection is often an early clinical sign and sometimes patients may also have hiccoughs, tachypnoea, bleeding, and shock. Dermatological indications include a typical maculapapular rash on trunk, but this is more frequently observed on white people than darker skinned people. Patients may also acquire neurological warning signs, namely, convulsions, delirium and coma. Patients normally die 6-9 weeks after the first indications. In surviving patients, improvement maybe gradual and is frequently illustrated by fatigue and arthralgia.50 From these clinical indications, it is clear that EVD may mimic several other tropical ailments like malaria, typhoid fever, or yellow fever at the start of the disease.
No FDA-approved vaccine or medicine (e.g., antiviral drug) is available for EBOLA. Symptoms of EBOLA and complications are treated as they appear. The basic interventions, when used early, can significantly improve the chances of survival such as, Providing intravenous fluids (IV) and balancing electrolytes (body salts); maintaining oxygen status and blood pressure; treating other infections if they occur. Experimental vaccines and treatments for EBOLA are under development, but many have not yet been fully tested for safety or effectiveness.
Isolation and infection control
Patients identified as being at risk of infection should immediately be isolated in a room with private bathroom facilities. All contaminated materials (such as clothes and bed linens) should be treated as potentially infectious. Specimens for laboratory investigations (such as EBOLA RT-PCR, full blood count, serum creatinine and urea, liver function tests, arterial blood gases, coagulation studies, blood cultures, and investigations for other conditions such as malaria) should be collected and sent off according to local and national protocols.
Fluid and electrolyte replacement
Vomiting and diarrhoea are common, so patients are often dehydrated and hypo volaemic, particularly if they present late. Oral rehydration solutions can be used for patients who can tolerate oral administration and who are not severely dehydrated. The volume of intravenous fluids needed should be assessed on the basis of the clinical examination (level of dehydration, signs of shock) and fluid losses (volume of diarrhoea or vomitus, or both). Large volumes of fluid replacement (>10 L/day) may be needed in febrile patients with diarrhea.33
Antivirals targeting viral entry step
Antibody-based EVD therapies have been studied from convalescent serum to monoclonal antibody and from single antibody treatment to antibody cocktail. In 1995, eight patients had symptoms of EVD, two of whom had even been in severe coma, and all of them were administered with convalescent sera from recovered EVD patients. Neutralizing antibody-based therapies can directly target the virion and cut off virus replication at the very early stage of viral entry, commonly used to treat post-exposure infection. Homogeneous polyclonal IgG purified from EBOLA vaccine immunized NHPs were also used to treat EBOLA- infected NHPs.55
A primary dose of 80 mg/kg of IgG at 48 h, and additional doses at 4 days and 8 days post-infection were provided to challenge NHPs. Besides, the potential antibody-dependent-enhancement of EBOV infection was reported previously.56 The antibody KZ52, derived from a survivor of the Kikwit ZEBOV outbreak in 1995, showed potent neutralizing activity at the cell culture level and was protective in a small animal challenge test under post-exposure conditions.57,58 The crystal structure of KZ52 and EBOLA GP trimer shows thatKZ52 recognizes the pre-fusion conformation of the GP trimer by clamping regions of the pre-fusion GP2 and part of GP1together.40
During past years, researchers have developed three generations of antibody cocktail formulations for EVD therapy. The first one was based on the combination of two human mouse chimeric mAbs, ch133 and ch226, which demonstrated strong neutralizing activity against ZEBOV in vitro. The second generation of anti-EBOLA antibody cocktail formulas, ZMAb and MB-003 consist of three completely different neutralizing mAbs derived from ZEBOVGP trimer antigen-immunized mice,49,50 ZMAb, containing mAbs 1H3, 2G4 and 4G7, showed 100%protection of cynomolgus macaques, with the first dose being given 24 h post-exposure followed by two additional doses every 3 days (25 mg/kg per dose). When the first treatment was administered 48 h post-infection, 50% of the cynomolgus macaques survived.59
The MB-003 cocktail, including antibodies c13C6, h-13F6, and c6D8, showed 67% protection in NHPs when treatment was initiated at 48 h post challenge with two additional doses (50 mg/kg per dose).60 The latest study tested different combinations of antibodies from MB-003 and ZMAb in NHPs challenge experiments, and the selected formulation with the best preventive effect termed ZMapp, was composed of c13C6 from MB-003 and two antibodies, 2G4 and 4G7, from ZMAb. All three mAbs recognized conformational epitopes located on GP2 or the stem region of the GP trimer, while the remaining three antibodies from MB-003 and ZMAb were bound to the trimer head. To increase the production of ZMapp, the compositional antibodies are currently produced from Nicotiana benthamiana.61 By applying a novel technology in plant protein expression system, named magnification, 500 mg of full-size monoclonal antibody can be purified from per kg fresh leaf biomass.62 This is an easier and cheaper approach to scale up the production of ZMapp, especially for developing countries. However, this technology may be insufficiently robust to scale up the production of ZMapp for combating the current EBOLA outbreak in West Africa.
Peptide-based inhibitor
The working principle of this kind of inhibitor is based on a CHR (or HR2) sequence containing peptide (C-peptide) that can interact with the NHR (orHR1) region in the viral GP2 and block the 6-HB formation between the viral GP2 NHR and CHR domains, resulting in inhibition of viral-host cell membrane fusion 42]. Unlike HIV, however, EBOLA viruses enter the target cell mainly through the endocytic pathway;39,40 thus, the peptide inhibitors must exist in the endosome where the 6-HB is formed. Hence, researchers added a HIV-1 Tat protein segment, which is known to target host endosome,63 to the N-terminal of EBOV CHR (610e633 AA of GP) with a short linker, and this synthesized peptide was called Tat-Elbow.64 Tat-Elbow as shown to enrich in endosome, and this location property proved to be associated with efficient entry inhibition of EBOLA pseudo virus and authentic EBOLA virus at cell culture level.
In another approach, CHR of GP2 was conjugated with cholesterol and a Tat analog to form a-helical conformation. In the EBOLA pseudo virus assay, infection could be reduced by1000-fold with this designed peptide at a concentration of 40 mM. On the other hand, the peptide could also inhibit the entry of control virus (VSV), suggesting its lack of specificity.65 Moreover, peptide entry inhibitors of EBOVs need to maximize the treatment window and minimize dosage.
Other approaches to interrupt the entry step
Several small-molecule compounds can also block cell entry of EBOVs. A high-throughput screening (HTS) of small-compound libraries was carried out based on a pseudo typed system, and a benzodiazepine derivative, termed compound 7, was identified to block the entry step of EBOLA virus and Marburg virus with an IC50 value of 10 mM and 12 mM, respectively. A kind of benzyl piperazine adamantine diamide-derived compound could inhibit the entry of EBOVs by preventing viral glycoproteins from binding NPC1.66 Similarly, inhibitors of CatL or CatB may also be antiviral candidates.67 Since drug candidates generally require several years of testing before approval for use in human, one research group screened licensed drugs to identify whether any of them would have anti-EBOV effects. They finally chose three clinically approved ion channel inhibitors, amiodarone (an ant arrhythmic drug), dronedarone and verapamil, all of which were previously shown to inhibit the entry of EBOVs based on pseudo virus assay results.
Small-molecules that interfere with RNA synthesis
Early studies showed that the commonly used anti-RNA virus drug, ribavirin could not limit the replication of EBOVs and failed to protect animals from lethal challenge.68,69 Recently, however, one pyrazine carboxamide derivative named favipiravir (T-705), which showed potent antiviral activity against numerous negative- or positive-strand RNA viruses.70-74 was demonstrated as an anti-EBOV drug in a mouse model.54,75 Favipiravir transforms into its active form, favipiravir-ribofuranosyl-5’-triphosphate (favipiravir-RTP), through phosphoribosylation by cellular enzymes in vivo. Favipiravir-RTP is considered to be a nucleotide analog that occupies the catalytic center of viral RdRP or incorporates into the newly synthesized viral RNA to cause lethal mutagenesis.76-78 Favipiravir inhibited RNA replication of ZEBOV in a cell culture model with an IC90 of 110 mM.
A prodrug of cidofovir, named brincidofovir (CMX001) showed potent anti-EBOLA activity at the cell culture level, and had been used in EVD patients’ treatment.79 Brincidofovir has been studied as the drug against several DNA virus infections,80 and is currently in Phase III clinical testing against cytomegalovirus and adenovirus.79 Brincidofovir inhibits viral replication by inhibiting viral DNA polymerases,81 so it may interfere with the RNA polymerase of EBOVs. Although the mechanism of anti-EBOLA activity is unclear, a new phase II clinical trials of brincidofovir was launched for testing its potential safety and antiviral activity in EVD patients.79 A novel adenosine analog, BCX4430, interferes with the function of RNA polymerase of EBOLA virus, and confers protection to EBOLA-challenged rodent animals.82
Oligonucleotide-based Antivirals
The technology of siRNAs was introduced to the anti-EBOLA field. The siRNAs specifically recognizing the RNA sequences of RdRP (EK-1), VP24 (VP-24-1160), and VP35 (VP-35-855) were identified, and they were packaged with polyethylenimine (PEI) or lipid particles for in vivo delivery.83 Another antisense oligonucleotide-based technology, termed phosphorodiamidatemorpholino oligomers (PMOs), was also applied for EVD therapy. This kind of DNA oligomers recognizes specific single-stranded RNA or DNA of viruses to form stable complexes in order to block viral replication.84 Although AVI-6002 and LNP/siRNA: TKM-EBOLA is under phase I clinical trials,85 two important issues affecting both approaches must be considered. For RNA viruses, the mutation rate at nucleic acid level is usually higher than the antigenic drift rate at protein level; therefore, antisense oligonucleotides-based drugs may face more problems in genetic variation of virus than other antivirals which target viral proteins like antibodies. Also both PMOs and siRNAs should be delivered into cytoplasm more efficiently to reduce the dosage and frequency of the drugs.
Drugs to modulate symptoms without directly targeting EBOVs
Since EBOVs antagonize the functions of type I interferon’s,86,87,88 exogenous interferon-a or interferon-b could delay the occurrence of viremia or prolong survival time, but not rescue NHPs from lethal infection.89,90 The coagulation disorders caused by EBOV infection are also an important factor in the development of EVD.91 Two licensed drugs the recombinant nematode anticoagulant protein c2 (rNAPc2)and the recombinant human activated protein C (rhAPC)originally used for anticoagulation, were approved to partially protect NHPs from ZEBOV lethal challenge, and the survival rates were 33% and 18%, respectively.92,93 However, these two approaches are unsuitable for use alone because of the irrelatively low efficacy, while they can be used as part of a cocktail treatment; and these drugs are much safer than newly discovered antivirals.
Post-exposure treatments
One of the EBOV prophylactic vaccines, an attenuated recombinant VSV expressing and displaying the GP of ZEBOV (VSV-EBOLA GP), has been studied as a therapeutic vaccine for EVD. This vaccine has not been approved for clinical trials, but in 2009, it was used in humans under an intractable situation. EBOVs are members of the so-called “viruses below rocks”, which have their own reservoirs and susceptible wildlife in the natural world. Human beings are susceptible, but only by intrusion into the life cycle of viral ecology in nature, meaning that this kind of virus cannot become extinct, like smallpox. For EBOVs, fruit bats and apes in Africa are considered as the reservoirs and susceptible hosts, respectively.94,95 By their culture, Africans normally come into contact with infected wildlife, and because of the underdeveloped system of public health in Africa, such high-risk contacts may develop into new outbreaks. Even worse, an animal challenge experiment indicated that inhaling aerosolized virus led to EBOV infections in NHPs.96 This means that EBOV may adapt to the respiratory transmission route and cause larger epidemics, or even pandemics.97 Therefore, the ideal plan would include rapid diagnostic methods, patient management, prophylactic vaccines, and post-exposure treatments in a combined effort to combat the disease. Once suspected cases have been confirmed, those infected and their close contacts must be isolated. Then, the local medical staff and inhabitants should be immunized with prophylactic vaccines immediately in order to prevent hospital-acquired infections and help break the chain of dissemination.
Actually, several viral vector-based or virus like particle-based prophylactic vaccines have exhibited 100% protection in EBOV challenge tests with NHPs.98,99 Since infected individuals may be discovered only after they have shown EVD symptoms, drugs with a relatively long treatment window, like ZMapp, are needed, and broad spectrum antivirals, like favipiravir, should be used jointly, since species-specific drugs may demonstrate low efficiency against variant strains or rare species of EBOV. However, almost all antivirals should be used at a high level of working concentration to provide complete protection, as mentioned above and could result in the demand for drugs exceeding the supply. Salutary lessons should be drawn from the anti-rabies strategy i.e., using therapeutic vaccines in order to activate acquired immune response against EBOVs. The utilization of both therapeutic vaccine and prophylactic vaccine would save drug resources in areas affected by EBOLA.
EBOLA viruses initially infect myeloid cells, which release numerous pro-inflammatory cytokines. These cytokines target endothelial cells, destabilize the actin cytoskeleton, and damage adherens and tight junctions, leading to a loss of endothelial barrier integrity, internal and external fluid losses, and vascular collapse.100
Since EBOVs are Biosafety level 4 pathogens, the facility limitation restricts the development of antivirals. To screen for antivirals that inhibit viral entry, EBOLA pseudo-typed systems based on either lentivirus backbone17 or vesicular stomatitis virus (VSV) backbone,18 which is conjugated with luciferase reporter gene, can be performed in BSL-2 laboratories.
Mini-genome replicon and partial reverse genetics systems, which can also be safely handled in BSL-2 laboratories, have been developed for screening chemical inhibitors to counter RNA transcription or
replication.20,101 For EVD symptoms caused by infection of EBOVs in primates, lethal challenge studies in NHPs are the gold standard for testing the effectiveness and safety of antivirals.24,61
To facilitate drug research, either a mouse adapted strain or an immune deficient mouse model has been used for preliminary testing.102,103 A similar strategy has also been applied in guinea pigs22 and Syrian hamsters.23 Normal test indicators for EBOLA animal models are survival rate, weight loss, body temperature; viremia, alanine amino transferases (ALT) and aspartame aminotransferase (AST) index.25
None.
None.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.