Journal of
eISSN: 2373-6453


The Malsoor virus (MV) is a novel Phlebovirus recently isolated from Rossetus bats from Western India that is closely related to Severe Fever and Thrombocytopenia virus (SFTS) and Heartland virus. Experimental studies showed pan cytotrophic property of the virus for different mammalian and insect cell lines in-vitro. High resolution transmission electron microscopy studies were carried out to characterize the fine structure and morphogenesis of this novel Phlebovirus. Detailed ultra structural studies on MV particles showed heterogeneous morphology. Majority of the virions (80%) were round having well defined envelope projections (6 ± 1nm) and having an average size of 70 nm. A subclass of virions (15%) showed pleomorphism. Interestingly, the envelope projections on the virions were tightly packed and not fragile to staining. Virus infected cells showed several interesting features not commonly reported for Bunya viruses. These included frequent detection of multiple inclusion bodies, distinct cytoplasmic membrane associated virus replication sites and presence of autophagy. This is the first electron microscopy study of this novel Phlebovirus. This is the first report on the detailed Ultra structure of this novel virus and forms the basis of further studies to explore the functional biology of this virus through more advanced approaches like correlative and cryo electron microscopy.
Keywords:Malsoor virus, Electron microscopy, Bats
MV, Malsoor Virus; SFTS, Severe Fever and Thrombocytopenia Virus; PID, Post Infection Day; PCR, Polymerase Chain Reaction; TEM, Transmission Electron Microscopy
Bunya viruses constitute a large group of viruses with potential as emerging zoonotic agents and these viruses have been isolated from different species ranging from bats to ticks. The family Bunyaviridae currently includes more than 350 RNA viruses that are distributed among five genera: Orthobunya-, Nairo-, Hanta-, Phlebo-, and Tospovirus.1 Genus Phlebovirus comprises over 70 antigenically distinct serotypes; the 68 known serotypes are divided into two groups: the Phlebotomus fever viruses (the sand-fly group, transmitted by Phlebotomine Sand flies) comprise 55 members and the Uukuniemi group (transmitted by ticks) comprises the remaining 13 members. Of these 68 serotypes, eight of them have been linked to disease in humans, e.g. Alenquer virus, Candiru virus, Chagres virus, Naples virus, Punta Toro virus, Rift Valley fever, Sicilian virus, and Toscana virus.2 Phleboviruses are geographically distributed in Europe, Africa, Central Asia, and the Americas and cause clinical syndromes ranging from a short, self-limiting febrile illness to fatal haemorrhagic fever.3,4
Very recently, during virus surveillance studies in wild bats in Western India, a novel Orthobunyavirus, the Malsoor virus (MV), was isolated from Rosettus species of bats.5 The virus showed broad host range infecting different cell lines and infant mice in-vitro. Importantly, this virus shows close genetic relatedness to two newly described and highly pathogenic human Phleboviruses, Heartland and Severe Fever with Thrombocytopenia virus (STFS).6,7 The Ultrastructure of Bunya virus morphogenesis and maturation remains incompletely understood and particle morphology have also shown variations. In the present study we examined these features of MV replication in the infected host cells.
Cell lines, virus and infection
The VeroCCL81cell-line was used for the present study. Cells were maintained in Dulbecco’s modified Eagle’s medium (GIBCO, USA) supplemented with 5% fetal bovine serum (GIBCO, USA) under 5% CO2 at 37oC. A prototype isolate of Malsoor virus (NIV 1050639) was used to infect confluent monolayer’s of the VeroCCL81 cells as described earlier.5 Cells were observed post-infection days (p.i.d) under inverted phase contrast microscope (Leica, Germany) for morphological changes.
Detection of virus replication by immuno fluorescence and polymerase chain reaction
Active virus replication in the infected cells was detected using an indirect immuno fluorescence assay (IFA) and polymerase chain reaction (PCR) with MV specific primers as described earlier.5
Negative staining and immuno electron microscopy
Supernatants from infected cell cultures showing distinct cytopathic effect and positive for MV RNA sequences by PCR were clarified and processed first for negative staining. Briefly, the culture supernatant was removed centrifuged at 4000g at 4oC for 10 minutes. The supernatant was harvested and subsequently centrifuged through a 30 % (wt/vol) sucrose cushion made in TEN buffers (0.01M Tris HCl pH 7.4, 1 mM EDTA and 0.1M NaCl) for 3 hr at 67,000 g under 4oC in an ultracentrifuge (Beckman, USA). The pellet was dispersed in 200 µl of TEN buffers and used for negative staining. Negative staining was done as described earlier.8 Briefly, a 50 µl aliquot of the virus suspension was adsorbed on a carbon coated formvar surface of a 400 mesh copper grid (Ted Pella USA) by floating for 20 minutes at room temperature. Excess fluid was removed and the specimen negative stained with 1% sodium phosphotungstic acid (PTA) pH 7.2. In separate staining, ammonium molybdate and uranyl acetate were also used to negative stain the MV particles.
For immuno electron microscopy, an aliquot of the MV suspension was mixed with 1:50 dilution of antisera against MV raised in mice and incubated for 45 minutes at room temperature. The mixture was then adsorbed on the EM copper grids and negative stained as described earlier.8
Transmission electron microscopy of cell cultures
Infected and control cells were harvested using a cell scrapper, pelleted by centrifugation and processed for embedding in EPON 812 as described earlier.9 Ultrathin sections were cut using glass knife in an ultra microtome (Ultra cut R, Leica Austria) and the 70-100 nm thin sections collected on 400 mesh copper grids. Staining was done using uranyl acetate and contrasted by Reynolds lead citrate. The sections were imaged under 120 KeV operating voltage in a Transmission Electron Microscope (TEM) (Tecnai 12, FEI, The Netherlands) and images were recorded with a side mounted with a 2K x 2K CCD camera (Mega view III, Olympus Germany).
Comparison of envelope glycoprotein sequences with other Bunya viruses
G1 and G2 amino acid sequences of each of the representative viruses were compiled using the Protein ID Feature on GenBank for corresponding M segment gene sequences. The G1 and G2 protein sequences were aligned separately using MEGA 6.0 software and CLUSTALW algorithm. Putative glycosylation sites for G1 and G2 proteins of each representative virus were identified with Expasy Prosite. The putative glycosylation sites were then mapped on the protein sequence alignments and compared with other Bunya viruses.
VeroCCL81cell line infected with the MV showed evidence of distinct cytopathic effect from the 2nd p.i.d. This was observed as rounding of cells with increased granularity and progressive detachment of the cell sheet (data not shown). IFA for MV antigens was positive by the 2nd p.i.d. Detection of MV nucleic acids was also positive by PCR from the 2nd p.i.d. (data not shown).
TEM examination of the negative stained MV particles showed several interesting observations. Majority of the virus particles were enveloped and showed a size distribution of 70 ± 3 nm as calculated from the TEM image analysis software (TIATM, FEI the Netherlands). The frequency of detection of different particle morphology classes after studying a significant number of particles (n=300) showed the presence of round particles with a dense envelope fringe as the most predominant class (approximately 80 %) (Figure 1a) followed by a pleomorphic form (approximately 15%) (Figure 1b) and a form that lacked distinct envelope morphology (approximately 5 %) (Figure 2a). This morphology was significantly rare. A schematic representation of the virion is shown in (Figure 1c). Immuno electron microscopy showed distinct clumping of the virus particles with an average of 4 particles per clump (Figure 2b). There were no significant differences in morphology of MV particles as seen with uranyl acetate and ammonium molybdate staining. The average projection length of the envelope was 6 ± 1nm and had dense packing where individual spike organization was not resolvable (Figure 1b-1c). In the MV particles where surface structures were clearly visible, penton-hexon symmetry was evident (Figure 2c).
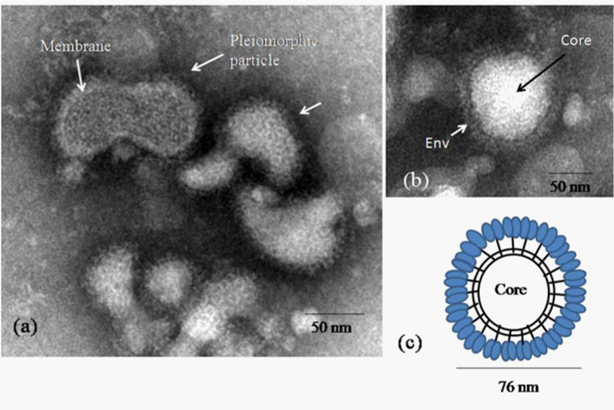
Figure 1 Negative stained morphology of the Malsoor virus
a)Representative negative stained electron micrographs showing variation in particle shapes in MV as seen in some fields
b)A typical enveloped virion
c)A schematic model of the complete virion of MV with envelope projections. Scales bars are built into the micrographs.
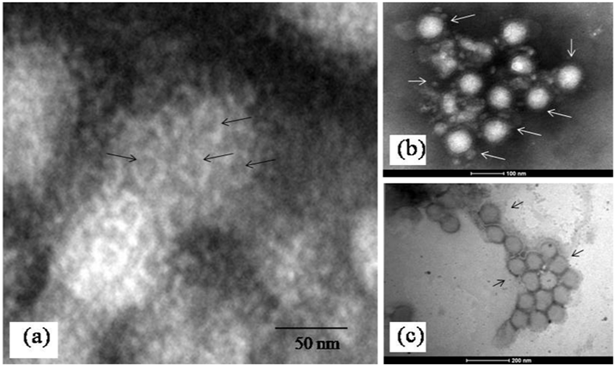
Figure 2Surface structure and IEM.
a)Surface projections seen as hollow cylindrical subunits on surface of an unfixed MV particle (arrows). Please refer to text for details b)A representative immune clump of MV particles c)Slightly deformed particles of unfixed MV particles showing relatively low surface contrast in negative staining. Magnification bars are built into the micrographs
The imaging of ultrathin sections of MV infected cells showed distinct changes. The most important of these was the presence of abundant cytoplasmic inclusion bodies. These structures often reported as virus replication factories or viroplasm have been described in host cells with several virus infections like pox, Rota and influenza but not commonly in bunya virus infected cells.10 In a recent paper similar cytoplasmic “viroplasm” or inclusion structures were reported in Vero cells expressing the non structural protein (NS) of SFTS virus in vitro.11 In the MV infected Vero cells, the inclusion bodies were mainly cytoplasmic. They were seen as rod shaped paracrystalline structures and electron dense crystalloid structures 400-500 nm in size (Figure 3a-3d & Figure 4a). The Ultrastructure of majority of the dying cells was suggestive of necrosis although an occasional apoptotic cell could be seen in the fields scanned. Interestingly, the evidence of autophagic engagement seen as formation of typical auto phagosomes and multi vesiculate structures was noticeable in approximately 25-30 % of the cells examined (Figure 4a-4d). Budding viruses could be imaged within Golgi compartments in multilayered vesicles (Figure 5a-5d). Extracellular virus particles were observed at moderate frequency. The Ultrastructure of majority of the dying cells was suggestive of necrosis while very few apoptotic bodies could be seen. Interestingly, the evidence of autophagic engagement was noticeable in approximately 25-30 % of the cells examined. This was seen as formation of typical auto phagosomes and multi vesiculate structures (Figure 4d-4e). Morphogenesis of MV was imaged associated closely with the Golgi complex. Extracellular virus particles were seen at moderate frequency and budding viruses imaged within Golgi compartments in multilayered vesicles (Figure 5).
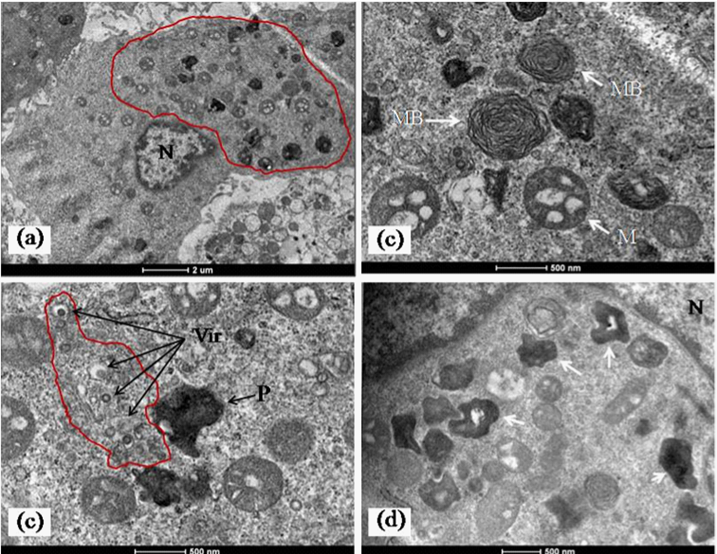
Figure 3 Representative images of inclusion bodies in MV infected cells.
a)A low power electron micrograph of an infected cell showing abundant cytoplasmic inclusion bodies without cytopathic effect in the cell. The area is encircled by the red contour line.
b)Typical cytoplasmic myelin bodies (white arrow, MB) and condensed mitochondria with electron dense matrix (white arrow, M) seen in abundance in most of the infected cells.
c)Areas of active virus replication associated with cytoplasmic endomembranes (arrow, Vir and area marked with red contour line. The field also shows a representative image of a electron dense crystalloid inclusion body (ref text for discussion) shown by arrow (black and P). A few scattered condensed mitochondria can be seen in the field.
d)An area of perinuclear crystalloid aggregates (white arrows). These were the most frequently detected inclusion bodies seen in the infected cells. Magnification scales are built into the micrographs.
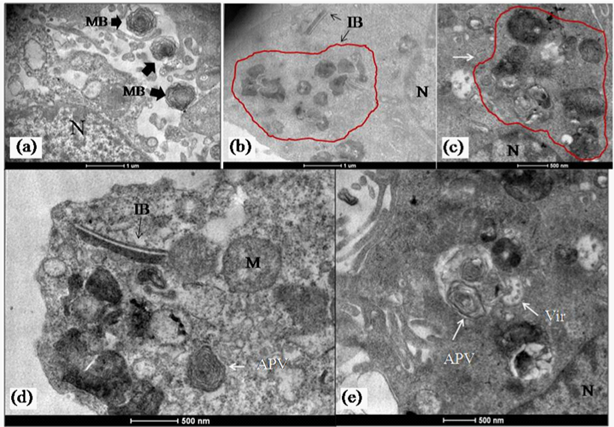
Figure 4 Presence of autophagy in MV infected cells.
a)(a-c,e) Representative image of MV infected cell showing presence of both inclusion body (IB, arrow) and areas of autophagic induction (arrow APV). An abnormal mitochondria (M) is also seen in (a,b).
b)A representative micrograph showing very typical ultra structural features of autophagic structures in the cytoplasm (arrow).Magnification bars are built into the micrographs.
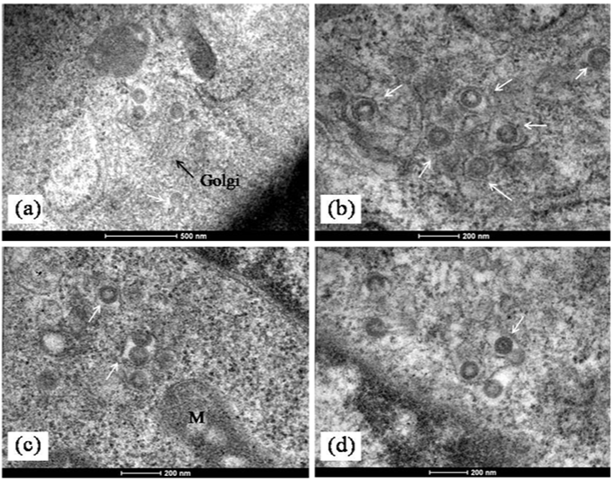
Figure 5 Morphogenesis of MV in infected cells.
a)Mature MV particles formed at the terminal end of a golgi stack (arrow). (b-d) Enveloped MV particles seen in dilated Golgi cisternae (white arrows). In
b)A complete virion with core is also visible (arrow). Scale bars are built into the micrographs.
G1 protein of Rift Valley Fever virus has one putative glycosylation site, not observed in other Bunya viruses (shown in yellow). First putative glycosylation site in the G2 protein of MV (yellow) also observed in SFTSV and Heartland Viruses (green) but lost in other Bunya viruses. Second putative glycosylation site in MV, also conserved in most other Bunya viruses (blue). The possible significance of these motifs playing a role in the virus assembly and structure is discussed in the text.
A notable observation from the sequence alignment studies of the G1 and G2 proteins coded by the M gene segment shows the presence of a N-linked glycosylation site at a conserved position between the Heartland, SFTS and MV (Figure 6). This site was seen to be present in Sand Fever Naples-like virus but lost in Rift Valley Fever virus. The family Bunyaviridae includes a very large number of genetically diverse, enveloped viruses having segmented RNA genome that is tripartite.12 While the biological properties and host range of these viruses are very diverse ranging from the rather ubiquitous endogenous agents of mosquitoes to viruses like La Crosse and Sin Nombre that cause severe disease in humans, Bunya viruses are surprisingly rather conserved in their overall structure.13 The Malsoor virus which was recently isolated from wild caught Rosettus bats showed very close similarity with the STFS and Heartland viruses at a genome level but the detailed structure and morphogenesis of STFS and/or Heartland viruses are not available to the best of our knowledge at the time of writing this paper. While both these viruses have been currently classified as high consequence emerging infectious disease agents, more studies are needed on MV before a confident classification can be proposed. In the present study we examined the detailed structure and morphogenesis pattern of MV in the host cell.
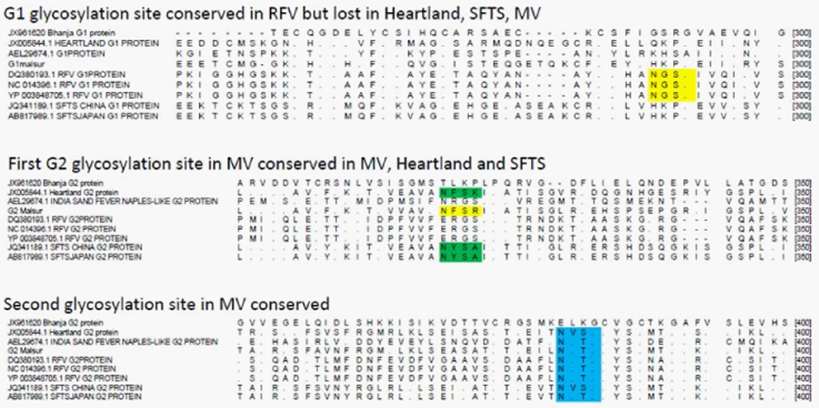
Figure 6 Alignment of G1 and G2 proteins of representative Bunya viruses showing putative glycosylation sites.
Using cryo electron microscopy (cryoEM) the 3D structure models of several Phleboviruses like Uukuniemi, RVFV14-16 and hantaviruses like Tula and Hantaan have been solved.17,18 Such studies have shown the multiple arrangements of Phlebovirus glycoprotein’s on the virion surface. Interestingly, since bunya viruses lack a matrix protein anchor base for the surface glycoproteins, the surface organization of the envelope glycoprotein’s Gn and Gc forms the crucial interaction for virion organization. The MV particles showed a surprisingly high degree of conservation of the envelope glycoprotein structure that was not fragile to negative staining. Interestingly, we could also detect morphological class variation in the cell free MV virions as described earlier for a Bunyamwera virus (V565 001 522) from infected BHK21 cells.19 Importantly, the observation of predominant occurrence of the enveloped particles of MV in cell free forms would argue towards a stable morphogenesis environment in the host cell. This was consistent with absence of any pleomorphic or shedded forms. In ultrathin sections the virus particles averaged 70 nm in diameter were predominantly round and showed distinct envelope projections. The maturing particles within Golgi vesicles showed a fine granular core structure.
The major site of virus assembly was seen to be associated with the Golgi and Golgi associated membrane compartments that are typical of Bunya virus assembly.20 However, an interesting observation was the detection of ultra structural changes suggestive of an autophagic process in a significant number of infected cells. Whether this is an outcome of cellular stress of infection in the host cell or directly mediated through fast replication of MV is speculative and not within the scope of the present and is being investigated separately. Another very interesting point that stands out from the present study is that although the overall morphology and morphogenesis events of Malsoor virus as imaged by transmission electron microscopy is consistent with the literature on Bunya virus replication, the virus was unusually small in size as compared with other Bunya viruses. This is consistent with an earlier report of another bat Bunya virus, Kaeng Khoi, isolated from Chaerephon plicata bats from Cambodia.21 Importantly, whether this has any association with the presence of N linked glycosylation sites seen shared with Heartland and STFS virus and could be a virulence marker is not clear. The public health importance of MV is remains incompletely understood and current studies are in progress to address this issue. This is the first report on the detailed Ultrastructure of this novel Phlebovirus and forms the basis of further studies to explore the functional biology of this virus through more advanced approaches like correlative and cryo electron microscopy approaches.
The fact that the Malsoor virus is closely related to the STFSV and Heartland viruses makes it imperative to study at the earliest all aspects of this agent and its impact on public health. Importantly, characterizing the morphology and fine structure of the virus is a major study as it provides significant clues towards understanding pathogenesis. What is unique from our study is the finding that the MV has certain unique morphological features compared to other Bunya viruses. These includes its relatively smaller size, rapid induction of cytoplasmic inclusion bodies, induction of autophagy and presence of a genetic motif related to the STFS and Heartland viruses. Importantly as this virus was isolated from bats the ecobiology and human exposure studies needs urgent studies.
The authors sincerely thank the staff Electron Microscopy & Pathology group and Mr Rajen Lakra, Technician, BSL4 laboratory for their dedicated support and technical assistance. Ms Nitali Tadkalkar is a graduate student with the EM group and a CSIR fellow. The work was supported through intramural funds from the Indian Council of Medical Research, Govt of India.
None.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.