Journal of
eISSN: 2574-9943


Research Article Volume 4 Issue 5
Correspondence: Audrey Wall, Nuritas, Joshua Dawson House, Ireland, Email wall.audrey@nuritas.com
Received: September 11, 2020 | Published: September 23, 2020
Citation: Kennedy K, Roi, Cal1, et al. pep_35E7UW, a natural peptide with cutaneous anti-ageing effects discovered within the Oryza Sativa proteome through machine learning. J Dermat Cosmetol . 2020;4(5):109-116 DOI: 10.15406/jdc.2020.04.00162
Background: The effects of both chronological and exogenous skin ageing have a profound impact on components of the extracellular matrix (ECM), leading to visible changes in appearance over time. Natural ingredients aimed at alleviating and counteracting the effects of cutaneous ageing are of great interest. Here, we investigate the anti-ageing potential ofpep_35E7UW, a peptide within the rice proteome identified by machine learning.
Aim: To examine the in vitro and ex vivo efficacy of pep_35E7UWon ECM neosynthesis in human dermal cells and skin explants.
Methods: Using machine learning, pep_35E7UW was identified and its anti-ageing effects were evaluated in human dermal fibroblasts (HDF’s) and keratinocytes (HaCaTs) by means of cell culture and wound healing assays. This was further assessed by real time qPCR, where the influence on key genes in the collagen and elastin synthesis pathways was examined. Finally, the ex vivo effect of pep_35E7UW was determined by immunostaining of human skin explants.
Results: Cell collagen and elastin assays of HDF’s treated with pep_35E7UWreported a 32% and 51% increase in each protein respectively. Gene expression analysis determined that pep_35E7UW significantly increased collagen and elastin expression at multiple time points following treatment. Cell migration was significantly increased with pep_35E7UW treatment in the wound healing assay. pep_35E7UW topical application resulted in significant increase in elastin expression (43%) in human skin explants.
Conclusion: Here, we demonstrate that pep_35E7UW, an artificial intelligence (AI) discovered peptide, has the capacity to be both an in vitro and ex vivo stimulator of fundamental ECM components. Our results position pep_35E7UWas a functional ingredient for attenuating the effects of cutaneous ageing.
Keywords: gene expression, bioactive peptide, natural, anti-ageing, in vitro, explants, machine learning
Cutaneous ageing is realised through two distinct processes, namely intrinsic and extrinsic ageing. Intrinsic ageing, which is regarded as an inexorable fate, is largely dictated, and ultimately predetermined, by genetics.1–3 Extrinsic ageing, however, is induced by environmental factors such as exposure to ultraviolet radiation (UVR), nicotine and air pollutants, and is therefore considered to be under a certain degree of voluntary control.4 The effects of intrinsic and extrinsic ageing have discrete manifestations; with chronological skin ageing typified by atrophy and finer wrinkles, and extrinsic ageing characterised by epidermal thickening, discoloration and deeper wrinkles.5 In spite of this, interestingly, the molecular pathways of intrinsic and extrinsic ageing largely converge,6–8 with the effects of UV induced photo ageing regarded as a superimposition on to those of intrinsic ageing.2,6,7,9
Chronological aging is typified by a loss of dermal collagen,10 with each passing year resulting in approximately a 1% decrease in the collagen content (per unit area) of skin.11 Impaired proliferative capacity of fibroblasts, reduced neosynthesis in the dermis and increased expression of degradative enzymes which target collagen, namely, matrix metalloproteinases (MMPs) all impact the effects of aging on collagen.1 Photoageing, associated with extrinsic ageing, can further impact on collagen in two distinct ways. Firstly, UV radiation leads to increased collagen breakdown by inciting the generation of reactive oxygen species (ROS) and the consequent induction of activator protein 1 (AP-1), which in turn elevates the downstream expression of MMPs.8,9,12,13 Furthermore, UV radiation can negatively impact on collagen synthesis by disrupting the expression of Transforming growth factor beta (TGF-β),12,14,15 an important cytokine involved in collagen production and fibroblast proliferation.12
While elastin only comprises 2-5% of the dry weight of skin,16 it is nevertheless imperative for conferring the properties of resilience17 and elasticity.10 As skin ages chronologically, a gradual loss of elasticity is experienced,1,16 with the onset of reduction in elastin gene expression occurring between the age of 40-50 years1. Histological alteration of elastin is considered a prime characteristic of extrinsic photoageing10, where unstructured elastotic material is accumulated in the dermis in response to UV exposure.4,10,18 Various MMPs have been shown to have a causative role in UV-induced elastinremodelling,16,19 and although the precise mechanisms of chronological alterations in elastin are not yet fully understood, MMPs are recognised as also playing a role.4,16,20
Treatment of ageing in skin is achieved through two broad mechanisms: protection from UV and approaches aimed at counteracting existing age damage.3,4,6 From the latter category, active ingredients designed to reverse the effects of ageing are ultimately concerned with inciting an effect on collagen, elastin or hyaluronic acid.4 However, in recent years, bioactive peptides have emerged as popular anti-ageing agents which have been proven to be capable of stimulating these major components of the extra cellular matrix (ECM).5,21,22
Computational approaches for bioactive peptide discovery have the potential to greatly accelerate the identification of novel functional ingredients.23 Artificial intelligence (AI and machine learning methods have been used recently to successfully discover a novel anti-inflammatory ingredient derived from rice24,25 and a natural peptide with anti-aging properties derived from the pea proteome.26 Here, we present in vitro and ex vivo evidence validating the efficacy of pep_35E7UW, a natural peptide identified in the proteome of Oryza sativa with anti-aging activity. Furthermore, we provide additional support to an in silico machine learning approach for discovery.
A machine learning approach was used to predict a natural peptide with anti-aging properties. In brief, pep_35E7UW was identified in the rice proteome, Oryza sativa, by a machine learning predictive architecture described previously by Kennedy et al. 202026. Briefly, unstructured data and structured data was curated for terms relating to anti-aging and the ECM. This data was processed by natural language processing (NLP) and graph-based (with manual curation) techniques respectively and formatted into a bespoke peptide representation to serve as the initial training set for the predictive model. The model was evaluated and trained with an independent test set of known functional peptides with the relevant bioactivity, and then this trained model was applied to a library composed of a distinct set of 7.6x106 plant sequences which were previously identified by LC-MS/MS. A maximum of three predict-test-refine loops were then executed, where the results of initial in house in vitro testing of peptides of interest which were predicted by the model to have ECM activity were subsequently integrated into the further training of the predictive machine learning model. A final prediction was carried out to identify pep_35E7UW from the proteome of O. sativa. To identify and filter out peptides exhibiting homology to know bioactive peptides BLASTP27 (word size=2; matrix=PAM30, e-value=10000) was used, which confirmedpep_35E7UW as a novel peptide.PEP_35E7UWwas produced by GenScript (Piscataway, NJ, U.S.A), where the peptide sequence and purity (95.5 -98.4 %) were validated by HPLC–MS/MS.
Cell culture
Primary human dermal fibroblasts (HDFs; normal adult skin) were purchased from Sigma-Aldrich (St Louis, MO, USA) and grown in 1g/L glucose Dulbecco’s modified Eagle’s medium (DMEM; Lonza Basel, Switzerland), supplemented with 10% foetal bovine serum (FBS; Bio-sciences Dublin, Ireland), 1% L-glutamine (Sigma-Aldrich, St Louis, MO, USA) and 1% penicillin–streptomycin (Sigma-Aldrich, St Louis, MO, USA). Cells were kept viable in culture in a humidified atmosphere of at 37°C, 5% CO2 until, at most, passage 16 was reached. Immortalised human keratinocytes (HaCaT) cells (Addex Bio, San Diego, CA, USA) were cultured in 4.5g L-1 DMEMat 37°C, 5% CO2 and supplemented with 10% (v/v) FBS, 1% (v/v) L-glutamine and 1% (v/v) penicillin–streptomycin.
Elastin and hydroxyproline ELISAs
To measure Elastin and Hydroxyproline (a surrogate marker for Collagen) protein expression, HDFs were firstly seeded in 24 well plates at 2×104 well-1densityand incubated overnight at 37˚C, 5% CO2. The following day, cells were treated with pep_35E7UWat 0.05µg/mL and 5µg/mLdiluted in DMEM supplemented with 10% FBS for a total of 72h. Supernatant and cells were then collected and lysed with cell lysis buffer (Cell Signaling Technology, Danvers, MA, USA) respectively, and used on quantitative ELISA kits to detect, elastin (sandwich ELISA; ELISA assays, Dublin, Ireland) and hydroxyproline (competitive ELISA; ELISAassays, Dublin, Ireland) protein expression following manufacturer instructions.
Extracellular matrix PCR array
To analyse the ECM gene expression, HDFs were plated at 6×104 well-1 density and incubated overnight at 37˚C, 5% CO2.The following day, HDFs were treated with 5μgmL-1 pep_35E7UW for 72h. Cells were then lysed with RLT buffer (Qiagen, Manchester, UK) and total RNA was extracted using the RN easy mini kit (Qiagen, Manchester, UK)as per manufacturer’s instructions. Total RNA (500ng) was reverse transcribed to cDNA using the RT² First Strand Kit (SABiosciences, Qiagen, Manchester UK Ltd). The synthesised cDNA was mixed with RT2 SYBR®Green qPCR Mastermix and then analysed on a 96 well RT2 Profiler PCR Array for human extracellular matrix and adhesion molecules (SABiosciences, Qiagen, Manchester UK Ltd) following the manufacturer’s guidelines. A 7900HT Real Time PCR System (Applied Biosystems, Warrington, United Kingdom) was used for the PCR analysis, and cycle threshold (CT) values were processed using the RT² PCR Data Analysis Software (SABiosciences, Qiagen, Manchester UK Ltd). The CT data for the 84 genes of interest were normalised using the geometric mean of the expression of housekeeping genes ACTB, 18s, GAPDH and RPLPO.
RNA isolation and quantitative PCR (qPCR)
HDFs were plated at 6×104 well-1 in duplicate in 24 well plates and incubated overnight at 37˚C, 5% CO2.The following day, cells were treated with 5μg mL-1 ofpep_35E7UW for 3h, 6h and 12h, after which cells were lysed with TRIzol (Invitrogen, Carlsbad, USA), duplicates pooled, and total RNA isolated according to the manufacturer’s instructions. Gene expression of elastin and collagen mRNA was assessed by qPCR using Taqman assays on demand Hs00355783_m1 and Hs01076777_m1(ABI Biosystems, CA, USA), respectively. β-actin was used as endogenous control gene (Hs99999903_m1). qPCR was performed with 1µl/well of reverse transcription products (1µg total RNA), obtained by using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher, Waltham, USA), in Taqman Gene Expression Master Mix (ABI Biosystems, CA, USA). A final volume of 6µl/well was loaded into a Micro Amp™ Optical 384-well reaction plate (ABI Biosystems, CA, USA) in duplicate and qPCR was performed on a ABI Quant Studio 7 Flex Real-Time PCR System (ABI Biosystems, CA, USA) real time thermal cycler. The threshold cycle (Ct) values were determined and normalised to endogenous control by using the instrument software and data analysis was based on the ΔΔCt method.
Immuno fluorescence staining
To stain the ECM proteins, HDFs were seeded at 3 × 104 well-1 and 1.5 × 104 well-1(for collagen and elastin staining respectively) on 18mm glass coverslips (Paul Marienfeld GmbH & Co. KG, Lauda-Königshofen, Germany), in a 24 well plate and incubated overnightat 37˚C, 5% CO2.. The following day HDFs were treated with 5μgmL-1pep_35E7UW for 24 h, after which they were fixed in 4% paraformaldehyde (Sigma, Arklow, Ireland). Subsequently, in brief, coverslips were washed, incubated with 0.1% Triton X-100 (Sigma, Arklow, Ireland) and blocked with 2% BSA solution (Sigma, Arklow, Ireland).Coverslips were then incubated with either mouse anti-human anti-collagen alpha 1 (COL1A1) monoclonal antibody (Novus Bio, Littleton, USA) or rabbit anti-human anti-elastin (ELN) polyclonal antibody (Abcam, Cambridge, UK), in a 1:200 dilution in 2% BSA overnight at 4°C. The day after, coverslips were washed and incubated with Alexa Fluor 488 goat anti-mouse IgG or Alexa Fluor 546 goat anti-rabbit IgG (Life Technologies, Eugene, USA), for COL1A1 and ELN staining respectively, in a 1:200 dilution in 2.5% BSA for 1h at RT. Coverslips were then washed again and mounted onto Superfrost Plus microscope slides (Thermo Scientific, Waltham, USA) using Vectashield Mounting Media for Fluorescence with DAPI (Vector Laboratories, Burlingame, USA). Fluorescent staining of COL1A1 or ELN was visualised at 200x using an Olympus Bx51 fluorescent microscope, and images were captured using a Nikon DSLR camera.
Wound healing assay
As described by Kennedy et al., 2020, HaCaT cells were seeded at a density of 6×104 well-1/24-well plate and cultured until they reached 100% confluence. Once the appropriate confluence level was reached, cells were serum-starved overnight. This was followed by the cell monolayer being scratched with a sterile 200μl pipette tip. Cells were then rinsed twice in Dulbecco's phosphate-buffered saline (DPBS; Lonza, Basel, Switzerland) and subsequently left untreated (as a control) or treated with pep_35E7UW (0.5μg mL-1). Phase contrast images were captured at immediately after wounding, at 24h and at 48h using an Olympus E-620 digital camera (Olympus Imaging Corp.) mounted on an inverted microscope (10x objective; Olympus CKX41). Relative migratory activity was measured by calculating the healed area using ImageJ software (Rasband, WS, US National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/, 1997–2017).
Human skin explants
NativeSkin®, a human skin explant model (surface area 0.5cm2) was obtained from Genoskin (Toulouse, France). The method used to obtain these explant models from human skin biopsies was fully described previouslyby Norsgaard et al.28 Culture and maintenance of explants was following manufacturer’s instructions. After arrival, before treatment, explants were firstly delipidated in 10µl alcohol for 10 min at RT. Explants were then treated with either pep_35E7UW (15μl of a 20uM solution) or autoclaved distilled water (control) twice daily for the following 5 days. After treatment, explants were formalin-fixed and paraffin-embedded for immunohistochemical localisation of Collagen 1A1 (ab138492, rabbit; Abcam Cambridge, MA, USA) and Elastin (ab77804, mouse; Abcam Cambridge, MA, USA) on a DAKO Link 48 Autostainer. Antigen retrieval (Agilent Dako PT LinK) was performed for 20min at 97°C in target retrieval buffer at pH 6 (elastin) and pH 9 (collagen). Primary antibodies were both incubated for 30 min at optimal dilutions, 1:5000 (collagen) and 1:500 (elastin) followed by appropriate linker (Agilent Dako Envision Flex Linker kit). The chromogen, Vector Nova Red (Vector Labs, Peterborough UK) was used for detection (15 min) and counterstained with haematoxylin. Negative and isotype controls were included. Whole stained slides were digitised at 20X using the AperioScan Scope XT digital scanner. The digital images were viewed with Image Scope software and annotated to analyse biomarker expression in the whole tissue. Staining intensity was quantified using a specific macro for each biomarker based on the Colour Deconvolution v9 algorithm (Aperio technologies, Vista, CA, USA).
Statistics
All data analysis and stats were performed by using R software.29 Data is shown as mean ± standard deviation (SD) and analysed by the Student’s t-test or ANOVA followed by Tukey’s test for post-hoc pair wise comparisons. Only p values< 0.05 were considered significant. Graphs were generated using the ‘ggplot2’ R package.30
Following a machine learning approach, a peptide sequence, IDGYDTPVEGR, was identified in the proteome of O. Sativa and predicted with anti-aging activity. pep_35E7UW is 11 amino acids long with a molecular weight of 1221.27 Da, has a global charge of -2 and an isoelectric point of 4.03.
Effects of pep_35E7UW on ECM proteins
Following prediction and synthesis, the influence of pep_35E7UW on collagen and elastin synthesis was assessed by means of separate quantitative ELISAs. After 72 hours of treatment with pep_35E7UW, human dermal fibroblasts demonstrate a significant increase in collagen expression compared to untreated cells (see Figure 1A). Elastin expression in human dermal fibroblasts is also significantly enhanced by pep_35E7UW compared to the untreated control, however, here a dose dependent effect is not observed (Figure 1B). The results of our cell elastin assay were further confirmed by fluorescent staining. Here, human dermal fibroblasts were treated with 5µg/mL of pep_35E7UWfor 24 h or remained untreated. Cells were then treated with a fluorescent elastin probe for detection of elastin synthesis. In Figure 1 C, we see that fibroblast cells treated with pep_35E7UWshow a substantial increase in elastin formation compared to untreated cells. These results support our cell-based assay findings that pep_35E7UWis a potent stimulator of elastin production.
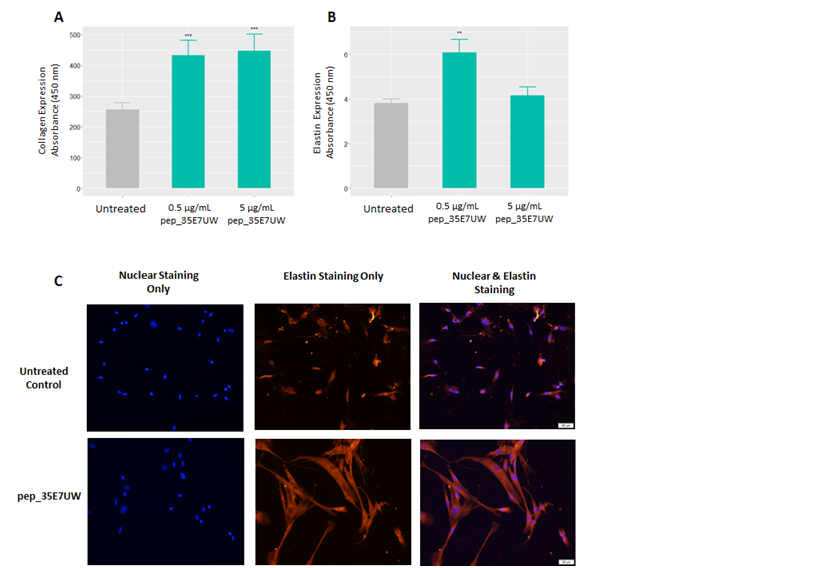
Figure 1 The effect of pep_35E7UWon collagen and elastin synthesis by human dermal fibroblasts. (A)Collagen and (B) Elastin assays were performed on HDFs treated with pep_35E7UW (0.5µg/mL& 5µg/mL) for 72 h. (C) Staining of HDFs treated with pep_35E7UW(5µg/mL) for 24h. Immunofluorescence for cell nuclei and elastin was achieved with DAPI and TRITC respectively. Images were taken at 200x magnification. (**p<0.01, ***p<0.001).
Gene expression profile of ECM and adhesion molecules following pep_35E7UW treatment
Using a real time PCR array, the effect of pep_35E7UW on the expression of 84 genes related to ECM maintenance and cellular adhesion were assessed in HDFs. Results were normalized to the expression of the β-actin housekeeping gene and depicted as fold change between fibroblasts treated with and without pep_35E7UW. The expression profile induced by treatment with pep_35E7UW for 72 hours is presented in Figure 2. Here, genes with a greater than or equal to 2-fold change in expression in response to pep_35E7UW are displayed.
Treatment with pep_35E7UW was found to alter the expression of 30 genes in dermal fibroblasts, where interestingly, all display up-regulation. In total, pep_35E7UW increased the expression of 11 adhesion molecules, including various integrins and laminin; 10 ECM proteins, including various collagens and fibronectin; and 9 molecules involved in ECM remodelling (Figure 2). Genes demonstrating a greater than 5-fold increase in expression in response to pep_35E7UW include collagens Type VII and VIII (COL7A1: 5.53; COL8A1: 8.54); integrin alpha-3/beta-1 (ITGA3: 5.45), a known receptor for various ECM components including collagen, laminin and fibronectin; and the metalloproteinases MMP-11 (5.55), MMP-14 (6.08) and ADAMTS13 (8.2).
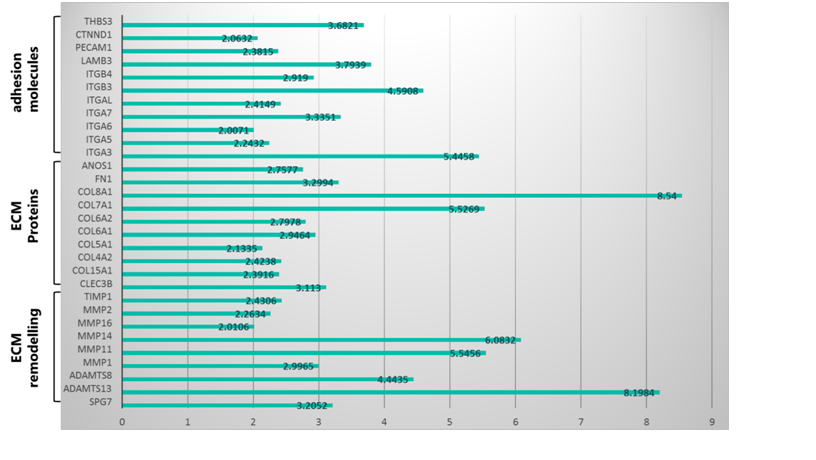
Figure 2 The gene expression profile of pep_35E7UW. The expression of ECM and adhesion genes was analysed via a PCR array using RNA from HDFs treated with 5µg/mL of pep_35E7UW for 72 h and expressed as fold change compared to untreated cells.
Quantitative PCR of select ECM proteins following pep_35E7UW treatment
qPCR experiments were performed for the following genes: COL1A1 and ELN. Similar to our array results, data were normalised by the β-actin housekeeping gene and expressed as fold over untreated control. As before, genes with a greater than or equal to 2-fold change in expression were considered.
Here, RNA from HDFs was extracted following 3, 6 & 12 h of treatment with pep_35E7UW (5µg/mL). In Fig. 3, we see a significant increase in collagen expression after 3 h & 6 h of treatment and a significant increase in elastin expression following 6h & 12 h of treatment. These results are in line with those reported in our array analysis, lending support for the functionality of pep_35E7UW as a promoter of ECM protein synthesis.
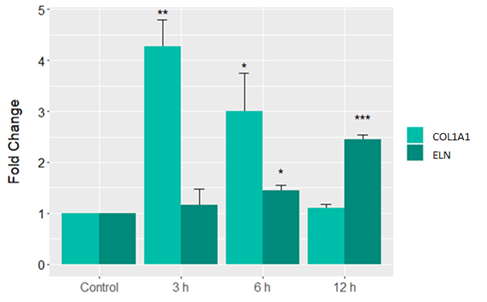
Figure 3 Gene expression profile of HDFS treated with pep_35E7UW for up to 12 hours. Time course analysis of elastin (ELN) and collagen (COL1A1) gene expression in HDFs treated with pep_35E7UW (5µg/mL) across 12 h. Fold changes are normalised to β-actin endogenous gene and relative to untreated HDFs at each time point. (*p<0.05 **p<0.01 ***p<0.001).
Effect of pep_35E7UW on wound healing
A scratch wound assay was performed on HaCaT cells to determine the wound healing properties of pep_35E7UW. The HaCaT cell monolayer was scratched with a sterile pipette tip, after which cells remained untreated or were treated with 0.5µg/mL of pep_35E7UW. Phase contrast images were captured after 48h and relative migratory activity was measured by calculating the wound area using image analysis software. In Figure4a, we see that pep_35E7UW considerably stimulates keratinocyte migration resulting in a reduced wound area compared to untreated cells. Quantification of the wound area demonstrates that pep_35E7UW significantly increases wound healing capacity 40% after 48h (Figure 4b).
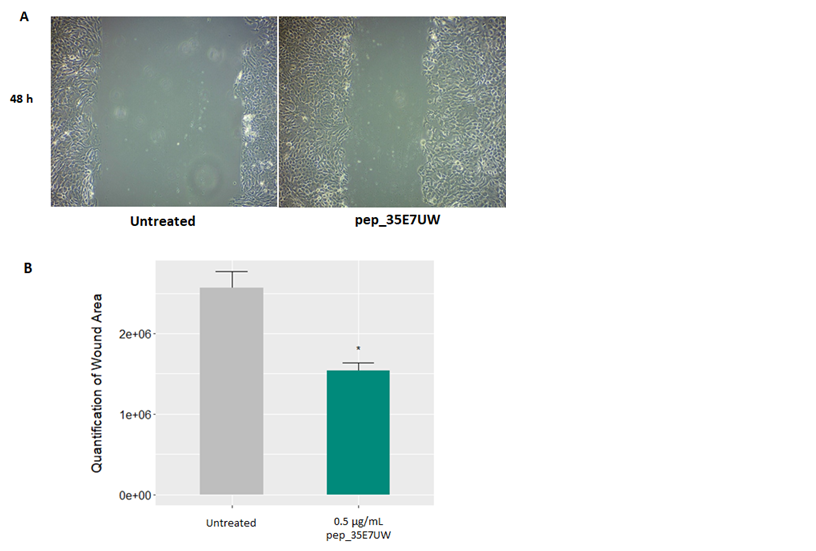
Figure 4 The effect of pep_35E7UW on human keratinocyte migration. (A)Phase contrast images of wound area in HaCat cells taken 48h after treatment with pep_35E7UW (0.5µg/mL) vs. untreated cells. (B) Quantification of the wound area as determined by imaging analysis. (*p<0.05).
Effect of pep_35E7UW on ECM protein stimulation ex vivo
Human explant models were topically treated with 20µM of pep_35E7UW twice daily for 5 days. Immuno histochemical analysis was then performed to investigate the collagen and elastin stimulatory responsestopep_35E7UW. Our elastin staining results are presented in Figure 5, where compared to the untreated control (Figure 5A), treatment with pep_35E7UW resulted in enhanced elastin staining (Figure 5B). Quantification of staining intensity by image analysis software revealed that pep_35E7UW significantly increased elastin expression by 43% (Figure 5C). Collagen staining results also demonstrated that pep_35E7UWhad a stimulatory effect compared to the untreated control, However, while image analysis quantification reported a 29% increase in collagen for explants treated withpep_35E7UW, this was not found to be significant (Figure 4D).
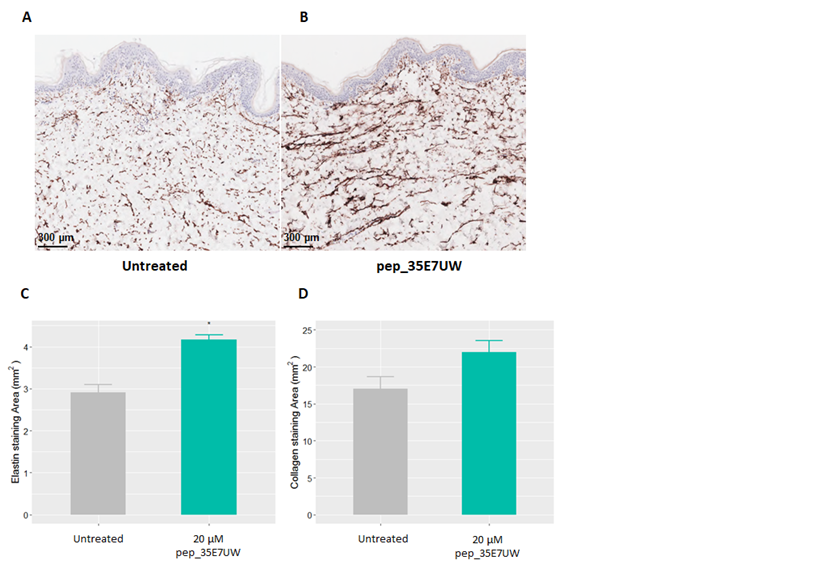
Figure 5 The effect of pep_35E7UWon elastin and collagen expression in human skin explants. Dermal morphology and expression of elastin in an ex vivo human skin explant model. Explants were treated with either water (A) (untreated) or (B) pep_35E7UW (5µl) twice daily. Expression of elastin was measured after 5 days of treatment. Morphometric quantification of (C) elastin expression and (D) collagen expression in the dermis at day 5 of pep_35E7UW treatment.(*p<0.05).
As high-throughput technologies and data space increase exponentially the amount of biological information available over the last few decades, computational approaches are being positioned as the most time-effective strategies to extract indecipherable knowledge and identify new peptide based ingredients and therapies in a growing number of health areas.31–33 Artificial Intelligence and machine learning approaches are increasingly seen as notable methodologies for discovery34with recent novel functional hydrolysates and peptides being described in the areas of inflammation and anti-aging.24–26 For example, Kennedy and Cal et al. Described for the first time the successful application of a deep learning approach for the discovery of a Pisum sativum (pea)-derived peptide with clinically proven functional skin anti-ageingproperties.26
O.sativa, the staple food for approximately 3.5 billion people,35 is a documented source of various functional anti-ageing ingredients36–38. Using a transferrable in silico approach, pep_35E7UW, a novel, natural peptide, was identified from the proteome of O. sativa. To our knowledge, this is the first time an anti-aging bioactive peptide derived from O. sativa and discovered using machine learning methods has been investigated. Here, through in vitro and ex vivo experiments we validate our computational prediction and presentpep_35E7UWas an effective stimulator of key ECM and epidermal components.
Evaluating our results, elastogenesis can be ascribed as the predominant activity of pep_35E7UW. Wrinkle formation in aging is promoted by the loss of the elastic fibre network in skin’s dermal layer.6,39 This is primarily due to the lower elastogenesis capacity of fibroblast coupled with and increased action of ECM degradative enzymes.40,41 Despite efforts in this specific area to date, elastin modulators are still not abundant or well characterised and is therefore a potential need and promising anti-ageing strategy to address.4,42 Here, we have shown that our peptide is seen to have a significant effect on elastin synthesis at the cellular level, increasing production by 60% which was confirmed through immunofluorescence; and at the gene level, inducing almost a 3-fold increase in elastin gene expression. Moreover, qPCR analysis of genes involved in elastin synthesis pathways exhibit expression profiles that align with that seen for elastin, providing evidence of widespread modulation by pep_35E7UW.
Reducedtionepidermal keratinocytes proliferation and migration of epidermal keratinocytes has also been previously linked to wrinkle formation in aging.43,44 Using a scratch wound assay, we assessed the effect of pep_35E7UW on keratinocyte cell migration. 48 h of healing resulted in a significant increase in cell migration in treated cells. Interestingly, as elastin is an essential functional component of the dermal ECM, it is also an important part of skin wound repair that encompasses an elastic dermis, and as such would be a favourable target in wound healing/scar reduction in mature skin.45 The gradual loss of elasticity in aging also leads to sagging skin, which is directly related to a slower wound healing and epidermal turnover. Potential therapies accelerating the cell cycle and the healing process will improve skin appearance and counteract sagging skin phenomenon.46
The most convincing evidence of the elastin stimulatory capacity of pep_35E7UW, however, is found in our experiments with human skin explants, where topicalapplication of our peptideis seen to significantly increaseelastin expression by 43%. Stimulation of elastin is recognised as a key strategy for the treatment of skin ageing4,5 yet remains under represented in terms of the functionality of activeingredients.47 While collectively, our results suggestpep_35E7UWholds great promise as a potentelastin stimulant, evaluation in a clinical setting is required to conclusively determine this functionality.
Production of collagen, also recognised asa fundamental approach for the treatment of skin ageing4,5,wasfound to be a further activity of pep_35E7UW.Collagen synthesis has been shown to be decreased in aged skin due, in part, to the potential lower collagen synthesis capacity o fold fibroblasts.48 Fortunately, this decrease may be reversible as compounds like retinoic acid have demonstrated the ability to stimulate and restore collagen production in aged skin.49 Our experiments with human dermal fibroblasts demonstrate that pep_35E7UW significantly enhanced collagen production by up to 75%, while PCR analyses reported a significant 4.2-fold increase in collagen expression. Using quantitative real-time PCR, collagen expression was found to be significantly increased at 3h and 6h. The transitory expression of collagen and difference kinetics to elastin expression observed here is understandable as different genes are known to exhibit different kinetics of expression.50 Although these results point to strong collagen stimulation in vitro, this failed to translate to a substantial effect ex vivo, where despite inducing a 29% increase in collagen expression, the effect of pep_35E7UWwas not found to be significant. Despite poorer performance in our skin explant models, the strongly suggestive in vitro collagen results still warrant further in vivo validation of pep_35E7UW.
Here, we present cogentevidence thatpep_35E7UWall eviates the effects of ageing in human skin by stimulating the production of key ECM proteins and epidermal keratinocytes. While, we have shown the efficacy of our peptide translates from an in vitro to ex vivo setting, translatability to anin vivo effect remains to be evaluated. Further studies are required to elucidate the clinical application of pep_35E7UW. Additionally, the results presented here, lend further support to an in silico machine learning approach for discovery.
None.
We declare no conflict of interest.
We thank Janet McCormack (Research Pathology Core, Conway Institute of Biomolecular and Biomedical Research, University College Dublin, Ireland) for technical assistance with immunohistochemical staining analyses. We also thank Neil Foster and Brian Keogh, both employees of Nuritas Limited, for useful discussion.

©2020 Kennedy, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.