Journal of
eISSN: 2378-3184


Research Article Volume 13 Issue 1
1Department of Marine Biology, Microbiology and Biochemistry, School of Marine Sciences, Cochin University of Science and Technology, India
2Assistant Professor, Research Department of Fisheries and Aquaculture, Conservation Research Group, St. Albert’s College (Autonomous), India
3Cochin University of Science and Technology, India
Correspondence: Radhika R, Department of Marine Biology, Microbiology and Biochemistry, School of Marine Sciences, Cochin University of Science and Technology, Cochin 682016, Kerala, India, Assistant Professor, Research Department of Fisheries and Aquaculture, St. Albert’s College (Autonomous), Kochi, Ernakulum, Kerala, India
Received: March 25, 2024 | Published: April 9, 2024
Citation: Radhika R, Santu KS, Bijoy NS, et al. Reconstructed phylogeny -glimpse of polyphyly of the micro-copepod genus Oncaea- (crustacea, copepoda: cyclopoida, family: oncaeidae) using mtCOI sequences substantiated by morphology. J Aquac Mar Biol. 2024;13(1):15-21 DOI: 10.15406/jamb.2024.13.00391
A fragment of the mtCOI gene was successfully PCR-amplified from two species (Oncaea venusta and Oncaea media; Family Oncaeidae; Cyclopoida) (534 to 640 base pairs) which represents the first molecular data. Objectives of this pioneering study were to derive their preliminary molecular phylogeny of based on mitochondrial (mtCOI) sequences; test if morpho-species are supported by molecular data. Reconstructed phylogeny postulated the Polyphyly of the genus Oncaea and paraphyly of its closely related genus Dioithona; Concluded the allopatric speciation and extended sovereign evolutionary history of O. venusta typica and O. venusta; Hypothesized the convergent evolution and homoplastic condition of the character (the absence of aberrant spine number in the distal segment of P1 endopod) in O. venusta; Re-confirmed the subspecies status of O. venusta typica to O. venusta through molecular study ; Hypothesized to retain the current genetic status of Oncaea sp. 7, O. parabathyalis, O. shmelevi and O. prendeli ; Recommended a detailed imminent morpho-molecular evaluation about the position of O. curta as the interspecific divergence values between O. waldemari and O. curta was found to be 2%.
Keywords: Lakshadweep, minicoy, oncaeidae, barcoding, Indian Ocean
Wilhelm Giesbrecht in his comprehensive monograph on the pelagic copepods of the Gulf of Naples established the family Oncaeidae.1 Literature data suggest that Oncaeids (genus Oncaea) have been reported from world’s major oceans and seas such as Mediterranean,2–7 the Red and Arabian seas,8–10 the NW and NE Pacific11,12 and the Antarctic.13 Redescribed O. mediterranea, O. clevei, O. media, O. scottodicarloi and O. waldemari from Korean waters.14 Many new species of Oncaea have been described from the Red Sea15,16 and many have been revised from the Mediterranean.17
The past few years have witnessed the increased recognition of taxonomic diversity of Oncaeidae cyclopoids - one of the most numerous members of the oceanic micro-copepod communities. While most of the existing reports and morphotaxonomic descriptions of Oncaeids are from Pacific (Korea, East Sea, Yellow Sea, China seas) and Atlantic Oceans (Adriatic sea, Mediterranean sea), the least has been accounted from the Indian Ocean except from Red Sea (but it is only an extension or inlet of the Indian Ocean. Since no taxonomic as well as molecular work on Oncaeids have been reported from the Lakshadweep Sea, this pioneering work stands significant in the major aspect of being a study from a biodiversity hotspot area (island ecosystems comprises 30% of the world’s biodiversity hotspots). Within the frame of a current taxonomic study from Lakshadweep Islands (South-western Indian Ocean), small Oncaeid cyclopoids (Genus Oncaea Philippi 1843) -Oncaea venusta and Oncaea media were morphologically identified from the samples taken from open ocean zones of Minicoy Island (Lakshadweep archipelago).
However, the dearth of molecular works in Oncaeids has certainly made it challenging to resolve various ambiguities related to their speciation. Never can a species be molecularly assessed unless and until it is morphologically identified. Blending the tools like DNA barcoding with conventional morphology has now constituted a powerful application for morphogenetic characterization of cyclopoid copepods.18
One of the precedence of this study was to reconstruct the phylogeny of the genus Oncaea and to test the generic polyphyly and paraphyly. Also, aimed to hypothesize the homoplastic condition of the absence of aberrant spine number in the distal segment of P1 endopod) in O. venusta. We also aimed to test if Oncaea sp. 7, O. parabathyalis, O. shmelevi and O. prendeli could be retained to their current genus “Oncaea” itself. Besides, we provide the primary barcodes of O. venusta and O. media based on mtCOI sequences that can be registered in the barcode catalogue of marine Oncaeid cyclopoids from South-Western Indian Ocean.
The samples subject to the present study were taken from eighteen offshore stations (open ocean) of Minicoy Island, Lakshadweep Archipelago, in April 2015 during the cruise (located between 8°12'-8°24'N and 72°54'-73°12'E) on board FORV Sagar Sampada Cruise #338 (Figure 1) where salinity was recorded as 33.5 to 34 PSU and temperature was in the range of 28 °C to 30 °C.
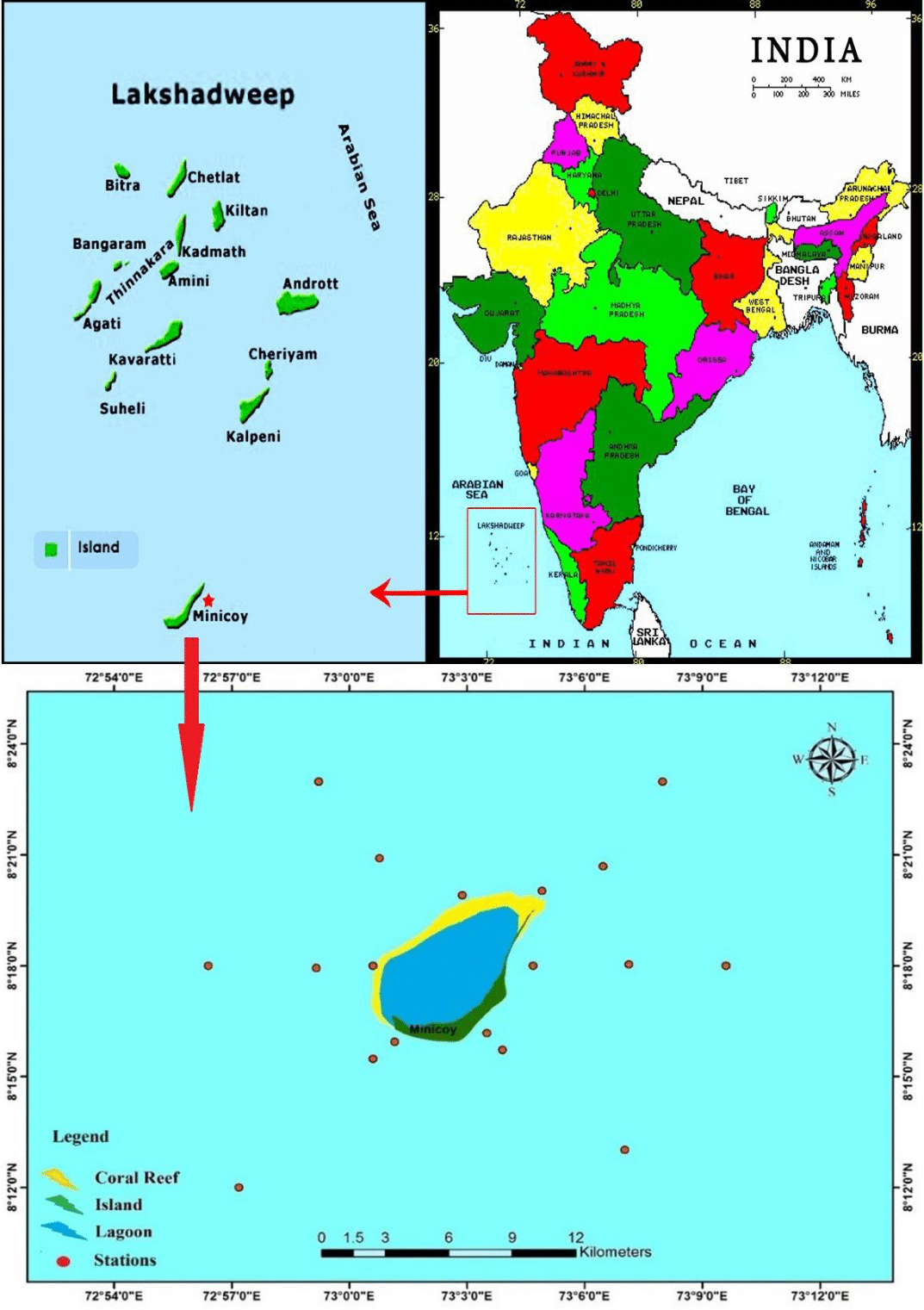
Figure 1 Study area- cruise track of FORV Sagar Sampada Cruise #338 (Minicoy Island), Lakshadweep, Indian Ocean.
For morphological and molecular analysis, the collected samples were then fixed in 4% buffered formalin and 95% ethyl alcohol respectively. Sorting, analysis and species level identification of Oncaea copepods were done under Stereo Microscope in the Ecology lab of the department using standard identification keys.14,16,17 Morphological examination and dissection of specimens were done in glycerol distilled water mixture using fine needles on a cavity slide under Leica (Model DM 500) bright-field compound microscope. Drawings were made with the aid of the drawing tube. Specimens were measured using ScopePhoto (x64) software. The specimens were incorporated into the copepod collection of Department of Marine Biology, Microbiology and Biochemistry, School of Marine Sciences, Cochin University of Science and Technology (catalog number MBM/DBT/27-28/2017).
Before DNA extraction, the alcohols preserved and were rehydrated in 500µl milliQ for 10-12hrs at room temperature.18 Individual copepods, cut into pieces with a needle, were transferred to a 2ml tube. Total DNA extraction was performed with the DNeasy Blood and Tissue Kit (Qiagen) using spin column protocol. The PCR reaction mixture contained 25µl PCR Master mix (Takara Clontech Emerald Amp® GT PCR Master Mix (Takara Bio, Otsu, Shiga Prefecture, Japan), 1 μL forward primer, 1μL reverse primer,8 μL template DNA, and 12 μL distilled deionised H2O.19 Reaction conditions included denaturation at 94°C for 1 min., annealing at 37°C for 2 min and extension at 72°C for 3 min which was carried out through 40 cycles. PCR products showing intense bands after agarose gels (1% with Ethidium bromide staining) electrophoresis were sent to ScieGenom labs (ScieGenom labs Pvt, Ltd, Kerala, India) for purification and sequencing. Obtained sequences were assembled using BioEdit 7.0.9 and aligned using Clustal X.20
Analysis was done using a final aligned length of 534-640 bp of the mtCOI sequences. The appropriate best-fit substitution model of DNA evolution was determined with ModelTest implemented in the software MEGA5 under the Akaike information criterion (A.I.C.). The mtCOI phylogeny analyses were conducted using the Tamura-Nei model with Gamma distribution. The rate variation among sites was modelled with a gamma distribution (shape parameter =1). The analysis involved 44 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All ambiguous positions were removed for each sequence pair. There was a total of 322 positions in the final dataset. Neighbor-Joining (NJ) method analysis was carried out to assess the relationships among the Oncaea, Oithona, and Dioithona species based on DNA sequence variation; relative support for the tree topology was obtained by bootstrapping using 1000 pseudoreplicates. Paracyclopina nana (Freshwater cyclopoid) was selected as an outgroup.21
Systematics
Family: Oncaeidae Giesbrecht, 1892
Genus: Oncaea Philippi, 1843
Species: Oncaea venusta Philippi, 1843
Specimens examined. One female dissected on one slide, one female in ethanol and two females destroyed for DNA sequences (Gen- Bank accession nos. KY368178.1, KY368179.1)
Female: Total length 1.27-1.38mm.Prosome 1.72 times as long as wide and 1.81 times longer urosome (Figure 2a & 3a). A1 six-segmented (Figure 3d).Urosome five-segmented (Figure 2b and 3b), 2.04 times as long as wide.GS 1.56 times as long as wide. CR 1.25 times as long as wide. Proportional lengths of the urosomal somites starting with 5th thoracic segment (+CR) is 0.9:7.6:0.3:0.3:1.2:2.
Distribution: Indian Ocean, SE Australia, New Zealand, Peru, Chile, Australia, East Korea, Cheju Island, China Seas, Malaysia (Sarawak: Bintulu coast), Mediterranean Sea ,Black Sea, Red Sea (http:// copepods. obs-banyuls.fr/en).
Family: Oncaeidae Giesbrecht, 1892
Genus: Oncaea Philippi, 1843
Species: Oncaea media Giesbrecht, 1891
Specimens examined: One female dissected on one slide, one female in ethanol and three males destroyed for DNA sequences (Gen- Bank accession nos. KT369529.1, KT369530.1, KT3695 31.1)
Male: Total length 0.911-0.95mm.Prosome (PR) thrice as long as wide as; 2.92 times longer than rest of the urosome (UR) (Figure 4a and 5a). The last segment of the prosome bears lateral lappets which are large, rounded with a sharp projection directed outwards. A1 four-segmented (Figure 5e).Urosome six-segmented, twice as long as wide.GS 1.31 times as long as wide as; longer than AS (Anal Segment) and CR combined. AS 1.17 times as long as wide.CR 2.94 times as long as wide (Figure 4 b,c). The first segment of maxilliped bears numerous spinous structures ventrally. Terminal claw 1.30 times longer than the preceding segment (Figure 5f).
Distribution: Epipelagic-mesopelagic. Inshore, coastal and oceanic, Gulf of Carpentaria, Great Barrier Reef, the North West Cape, New South Wales and south east including Tasmania, Pacific, Indian and Atlantic Oceans (http:// copepods. obs-banyuls.fr/en).
Further than eight Oncaeids species morphologically identified, only two species (O. venusta and O. media) could be successfully amplified using mtCOI sequences (average sequence length of 534 to 640 base pairs). Dioithona rigida (5 sequences) which is produced during the present study18 are also included in the analysis to demarcate Oithona and Dioithona species. There was no evidence of stop codons, ambiguities or insertions-deletions indicative of non-functional copies of mtCOI during the translation process of all sequences to protein using MEGA software. BLAST analyses of Genbank publicized that the developed sequences are copepod in origin and not contaminants, and 33 of the Genbank COI sequences (Table 1) and one of the sequence from the species Paracyclopina nana (KP 899609.1) as the out group.
|
S.No |
Species |
Region |
Accession number |
Remarks |
|
1 |
Oncaea venusta |
Lakshadweep (Lak) |
KY368178.1, KY368179.1 |
Developed |
|
2 |
Oncaea venusta typica |
Mediterranea (Med) |
|
NCBI |
|
3 |
Oncaea media |
Lakshadweep (Lak) |
KT369530.1, KT369531.1, KT369529.1 |
Developed |
|
4 |
Oncaea curta |
Gulf of Naples (GoN) |
KX650376.1 |
NCBI |
|
5 |
Oncaea mediterranea |
Mediterranea (Med) |
AB457134.1 |
NCBI |
|
6 |
Oncaea cf. mediterranea |
Mediterranea (Med) |
AB457130.1 |
NCBI |
|
7 |
Oncaea parabathyalis |
Mediterranea (Med) |
AB457147.1 |
NCBI |
|
8 |
Oncaea prendeli |
Mediterranea (Med) |
AB457146.1 |
NCBI |
|
9 |
Oncaea scottodicarloi |
Mediterranea (Med) |
AB457132.1 |
NCBI |
|
10 |
Oncaea scottodicarloi |
Gulf of Naples (GoN) |
KX650375.1 |
NCBI |
|
11 |
Oncaea shmelevi |
Mediterranea (Med) |
AB457145.1 |
NCBI |
|
12 |
Oncaea waldemari |
Mediterranea (Med) |
AB457136.1 |
NCBI |
|
13 |
Oncaea sp.7 |
Mediterranea (Med) |
AB457138.1, AB457139.1 |
NCBI |
|
14 |
Dioithona rigida |
Lakshadweep (Lak) |
KR528587.1, KR528586.1, KP972541.1, KP972540.1, KR528588.1 |
Developed |
|
15 |
Oithona dissimilis |
Japan, Okinawa Katabaru beach) |
AB604159.1 |
|
|
|
|
Japan:Okinawa, Hija-gawa River |
AB604161.1, AB604162.1, AB604164.1, AB604163.1 |
NCBI |
|
16 |
Oithona nana |
Sognefjorden Fjord |
AF259667.1 |
NCBI |
|
17 |
Oithona similis |
Korea |
JF269186.1, JF269185.1, |
|
|
18 |
Oithona simplex |
Kaneohe Bay |
KC594151.1, KC594150.1, KC594149.1, KC594148.1, KC594147.1, KC594146.1, KC594145.1 |
NCBI |
|
19 |
Oithona oculata |
Kaneohe Bay |
KC594144.1, KC594143.1, KC594142.1, KC594141.1 |
NCBI |
|
20 |
Oithona attenuata |
Kaneohe Bay |
KC594140.1, KC594139.1 |
NCBI |
|
21 |
Paracyclopina nana |
|
KP899609.1 |
NCBI |
Table 1 List of all sequences used in phylogenetic analysis
Extravagant average pairwise distances were observed between morpho-species (Table 2) with the lowest divergence (2%) between O. waldemari and O. curta; (18 %) between Mediterranean O. mediterranea and O. cf. mediterranea; O. parabathyalis and Oncaea sp. (22%); O. scottodicarloi and O. curta (23%). While the other taxa had average pairwise distances in excess of 23% which indicates the distinctiveness of each species.
Oncaea genus resulted polyphyletic in our analysis with 9 species assembling in one robust clade and sequences of O. venusta clustered in separate clade with Oithona similis. Another incongruence was that the sequence of Mediterranean O. venusta typica clustered with Oithona similis rather than with Lakshadweep O. venusta which is evidently indicated by their average pairwise distances (43%).
The NJ analyses (Figure 6) supported the presence of not less than 16 divergent lineages with high bootstrap values (>94%). A sister group relationship of O. media to O. scottodicarloi and O. waldemari /O. curta was supported by a high basal node bootstrap value of 87%. Another relative assemblage according to speciation was exhibited by O.mediterranea and O. cf. mediterranea. However, a moderately supported lineage (33%) which united three species: Oithona similis, O. venusta typica and O. venusta could be due to the low phylogenetic resolution of the mtCOI gene in basal nodes of the trees and perchance due to the saturation at third codon positions22 and also by various lengths of the fragments amplified. Another moderately supported lineage (39%) was of O. shmelevi, O. parabathyalis and Oncaea sp.7 next to O. mediterranea. There is also a moderately supported sister group relationship of Oithona (Dioithona) oculata and Dioithona rigida (bootstrap support 40%).
An indication of Polyphyly of genus Oncaea turned out to be the vital verdict in our COI phylogeny. Allopatric speciation and extended sovereign evolutionary history of O.venusta typica and O.venusta was concluded. Even though, the assemblage of Mediterranean O. venusta typica and Lakshadweep O. venusta with Oithona similis rather than with other Oncaea species was quite embarrassing; the status of O. venusta typica being the subspecies of O. venusta was quite re-confirmed through molecular study. Also, hypothesize to retain the current genetic status of Oncaea sp. 7, O. parabathyalis, O. shmelevi and O. prendeli.
Sewell (1947) identified O. venusta typica (0.75-1.23mm) as subspecies of O. venusta. In our analyses, the sequences of Mediterranean O.venusta typica and Lakshadweep O.venusta assembled with O. similis with a comparatively high interspecific distance (43%) suggesting their allopatric speciation (Mediterranean vs Lakshadweep) and long sovereign evolutionary history which in turn is mirrored in their numerous morphological differences. ie. The foremost character that presently abetted to distinguish O. venusta (Lak) is the absence of aberrant spine number in the distal segment of P1 endopod (Figure 3f) in addition to the pattern of sclerotization. The position of O. venusta and O.venusta typica deep inside the Oithonids group suggests that this distinguishing character might have originated independently. The absence of aberrant spine number in the distal segment of P1 endopod in O. venusta could be a homoplastic that might have evolved convergently.
The lowest divergence (2%) perceived between two allopatric species: O. waldemari and O. curta is questionable as the threshold value on intraspecific divergence is 0-4%.18,23 Meanwhile, O. waldemari from the Mediterranean Sea has been redescribed successfully with molecular support;24 probably, O. curta (GoN) could be a misidentification which needs a detailed morpho-molecular evaluation as suggested by the authors themselves.
The low divergence value of (18%) between Mediterranean O. mediterranea and O. cf. mediterranea; (22%) between O. parabathyalis and Oncaea sp.; (23%) between O. scottodicarloi and O. curta (probably O. waldemari) (Table 2) hint at either the higher speciation rate or slow evolution of this gene.22
A relatively very high average divergence values between any other sympatric Oncaeid and Oithonid species (all in excess of 23 %) implies a long sovereign evolutionary history echoed in their copious morphological dissimilarities. All these denote a niche segregation with trifling resource competition that has been observed and proved in several other Australian harpacticoid copepods.22
A reallocation of Oncaea sp. 7, O. parabathyalis, O. shmelevi and O. prendeli into another genus24 who proposed a monophyly of Oncaea genus in their phylogeny study where the aforesaid species did not confine to the monophyletic group of Oncaea. However, in our analysis, Oncaea sp. 7, O. parabathyalis, O. shmelevi and O. prendeli did restrain to the robust clade on Oncaea species (Figure 6) and proved their sisterhood relationship which is significant. Therefore, our study assumes to retain the current genetic status of Oncaea sp. 7, O. parabathyalis, O. shmelevi and O. prendeli, not a reallocation.
Morphology, however, did suggest that the two Lakshadweep Oncaeids are quite similar in comparison to previous descriptions14,25–28 and testing that hypothesis was one of the major aims of our study. Despite each species being represented with two (O. venusta) and three (O. media) sequences, interspecific divergences with other Oncaea sp. and Oithona sp. were much higher which is in complete accordance with previously observed morphological evidence.
Postulated the polyphyly of the genus Oncaea and paraphyly of its closely related genus Dioithona; Concluded the allopatric speciation and extended sovereign evolutionary history of O.venusta typica and O. venusta; Re-confirmed the subspecies status of O. venusta typica to O. venusta through molecular study and hypothesized to retain the current genetic status of Oncaea sp. 7, O. parabathyalis, O. shmelevi and O. prendeli.
Finally we propose 1) Sequence mtCOI from as many as Oncaeids from different regions of the Indian Ocean and compare the genetic congruence/incongruence 2) a detailed morpho-molecular evaluation about the position of O. curta as the interspecific divergence values between O. waldemari and O. curta was found to be 2%.
The authors owe their gratitude to Head of the Department of Marine Biology, Microbiology and Biochemistry, School of Marine Sciences, Cochin University of Science and Technology for the facilities and the Department of Science and Technology and Union Territory of Lakshadweep for research permission and other logistic support and necessary facilities for undertaking the study. This study forms a part of a major project “Taxonomy and genetic characterization of pelagic copepods (Crustacea) from the marine habitats of the South West Coast of India” funded by Department of Biotechnology, Government of India [BT/PR4258/AAQ/3/575/2011].
This work was supported by Department of Biotechnology, Government of India [BT/PR4258/AAQ/3/575/2011] of a major project “Taxonomy and genetic characterization of pelagic copepods (Crustacea) from the marine habitats of the South West Coast of India”
The authors declare that there are no conflicts of interest.

©2024 Radhika, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.