Journal of
eISSN: 2572-8466


Review Article Volume 7 Issue 5
Department of Natural Products, Institute of Pharmaceuticals Technology, Brazil
Correspondence: Raquel Elisa da Silva-López, Researcher of Department of Natural Products, Institut of Pharmaceuticals Technology, Fiocruz, Rio de Janeiro, Brazil
Received: July 25, 2020 | Published: September 9, 2020
Citation: Silva-López RE. The main protease of SARS-CoV-2 as therapeutic target to development specific drugs to treat COVID-19. J Appl Biotechnol Bioeng. 2020;7(5):185-189. DOI: 10.15406/jabb.2020.07.00232
The pandemic disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) denominated COVID-19 is an important problem of world public health because do not have specific drugs and vaccines available to control the disease progression and the rapid virusspreading. SARS-CoV-2 genome resemble to other SARS-CoV andMERS-COVcoronaviruses genomes, thus drugs developed for SARS and MERS treatment may be used for COVID-19. The aim of this work is discuss the molecular target of SARS-CoV-2, the main protease, a chymotrypsin-like named as 3CLpro, for a rational devolpment of specific molecules anti-SARS-CoV-2 with broad-spectrum anti-coronavirus activities.This study is a literature mini review using 47 articles obtained from Public Medline and Science directmainly from this year 2020. The coronaviruses infection is mediated by spike protein from viral capsid which bind to ACE2 receptor in the host cells which endocytes the viruses RNA, which is translated to proteins that are cleavead by proteases, mainly the 3CLpro. This enzyme is crucial to SARS-CoV-2 replication and its inhibition can prevent the virus replication. Thus, it is an important coronaviruses target because the amino acid sequence of 3CLpros from SARS, MERS and SARS-CoV-2 have high similarity. In silico studies have been demonstrated that synthetic small molecules, natural products, peptidomimetic inhibitors, and HIV inhibitors had different docking scores with SARS-CoV-23CLpro. However, peptidomimetic inhibitors seems to be better inhibitors to this protease and promise dugs to treat COVID-19 and other diseases caused by coronaviruses.
Keywords: SARS-CoV-2, Covid-19, therapeuctic target, 3CLproMain protease, inhibitors
E, envelope; M, membrane; N, nucleocapsid, S, spike
On December 31, 2019, chinese health authorities alerted the World Health Organization (WHO) to several cases of pneumonia without a recognized aetiologyin Wuhan City, Hubei province, China. The first cases were reported on December 8, 2019 and on March, 11, 2020 COVID-19 gained pandemic status.1 This pathogen was denominated as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the Coronavirus Study Group and the disease was named coronavirus disease 2019 (COVID-19) by the WHO.2 COVID-19 is affecting 217 countries and territories around the world and 2 international conveyances with more than 9.1 million confirmed cases and about 470 thousand confirmed deaths.1 Billions of people have been impacted due to mandatory lockdowns, isolations, and quarantines. Thus, the severe effect of the COVID-19 outbreak has imposed major challenges for global health, society and economy. Nowadays, the COVID-19 treatment and control are very difficult because there are no specific antiviral drugs or vaccines available. The prevention and supportive therapies are being implemented to avoid new cases and minimize clinical complications.3
Coronavirus
The current classification of coronaviruses recognizes 39 species in 27 subgenera, 5 genera and 2 subfamilies that belong to the family Coronaviridae, suborder Cornidovirineae, order Nidovirales and realm Riboviria. Coronaviridae are enveloped single-stranded ribonucleic acid (RNA) viruses that infect humans and animals. The size of positive-sense RNA genome of CoVsis between 26.2 and 31.7 kb.The spike projections of these virions give the appearance of solar corona. The main structural proteins of these viruses are envelope (E), membrane (M), nucleocapsid (N), and spike (S) (Figure 1). Coronaviruses outbreak has been reported three times in the earlier of 21st century namely SARS in 2002 (severe acute respiratory syndrome caused by SARS-CoV), MERS in 2012 (Middle East respiratory syndrome caused by MERS-COV), and the most recent COVID-19, caused by the new coronavirus, SARS- CoV-2.2 Seven type of coronaviruses cause human diseases. The four of them (HCoV-NL63, HCoV-229E, HCoV-OC43 and HKU1) induce only mild respiratory diseases, typically only in upper respiratory tract although some of them can cause severe infections in infants, young children, and elderly individuals.4 The two highly pathogenic and extremely invasive viruses, SARS-CoV and MERS-CoV, cause severe respiratory syndrome in humans and have high mortality rate, about 11 and 35 % for SARS-CoV and MERS-CoV, respectively.5,6 The transmission rate of SARS-CoV-2 is much higher than SARS-CoV and MERS-Cov and other flu-viruses.7 The clinical course of SARS and MERS was highly similar, and they also have similar pathogenesis.8 The coronaviruses attack patient's lower respiratory system by invading the airway epithelial cells, parabronchial epithelial cells, alveolar epithelial cells, vascular endothelial cells and macrophages in the lung. The pathological changes include focal haemorrhagic necrotising pneumonia with exudative diffuse alveolar damage,which can be fatal. They also affect gastrointestinal system, kidney, liver, heart, and central nervous system.5–9
Importantly, SARS-CoV-2 shared approximately 80% of the genome (~30,000 bases) with SARS-CoV and almost all their encodedproteins are homologous and also shows some similarities to MERS-CoV genome.10Thus, it will be very important to study the relationship among SARS-CoV, MERS-CoV, and SARS-CoV-2 for developing broad-spectrum antiviral therapies. Accordingly, drugs and other therapies for treating SARS may be used as a reference for COVID-19 treatment. The viral lifecycle steps provide potential targets for drug therapy.11 Promising drug targets include nonstructural viral proteins because their sequence are found in non-variable region of RNA, such as main 3-chymotryps in-like protease (3CLpro), papain-like protease(PLpro), RNA-dependent RNA polymerase, which share homology with other novel coronaviruses.12,13 Additional drug targets include viral entry, and immune regulationpathways.13,14
Infection and replication
The first step of viral lifecycle is the infection through the binding to a host cell receptor. The glycoprotein spike (S) on the outer surface of coronaviruses and the host transmembraneserine protease 2(TMPRSS2) are responsible for the attachmentto the angiotensin-converting enzyme 2 (ACE2)receptor andentry into the cell targets.15 This serine protease hydrolyzes specific peptide bonds in S protein and expose the ACE2receptor-bindingdomain.16 Furin, other transmembrane serine protease, can also be associated to ACE2 receptor, cleaves the S protein and facilitate the entry of SARS-CoV-2 in host cell after binding.17 Lung cells, enterocytes, nasal secretory cells co-express high levels of ACE2 and TMPRSS2.18 SARS-CoV infection reduces ACE2 expression in lung cells. Becauseloss of pulmonary ACE2 function is associated with acutelung injury, virus-induced ACE2 downregulation maybe important for disease pathology. ACE2 regulates the renin–angiotensin system (RAS). Therefore, a reduction in ACE2 function after viralinfection could result in a dysfunction of the RAS, whichinfluences blood pressure and fluid/electrolyte balance,and enhance inflammation and vascular permeability in the airways.18,19 Following receptor binding, the virus particle uses host cell receptors andendosomes to enter cells. SARS-CoV-2 deliver its nucleocapsid and hijacking the cellular machinery to replicate in the cytoplasm.The RNA genome composes of six to ten open reading frames (ORFs). The virus synthesizes RNA via its RNA-dependent RNA polymerase and encodes two ORFs, ORF1a, that is the longest part of the RNA encodes for the replicases, and ORF1b, which expresses for two large replicative polyproteins (pp) including pp1a and pp1ab comprising about 4000 (~500 kDa) and 7000 (~800 kDa) amino acids.20T hese polyproteins are auto catalytically processed to produce two proteases: a chymotrypsin-like proteaseserine protease (3CLpro Main Protease or 3CLpro) and a papain-cysteine proteinases (PLpro). These polyproteins are further processed to generatefour structural proteins and sixteen non-structural proteins (nsps). The proteolytic processing of the sixteennsps by 3CLproandPLpro is essential for virus replication and maturation because the nsps are involved in downstream binding and replication events including the formation of the replicase complex, which is essential for viral replication and transcription of the genome.21,22 The 3CLpro (33.8 kDa) cleaves the poly protein 1abat 12 distinct sitesin most of peptide bonds involving Leu, Gln↓ (Ser,Ala,Gly-↓ marks the cleavage site)to generate nsps4 to 16 which are essential for viral replication, because they perform function as RNA dependent RNA polymerase, RNA binding proteins, exoribonuclease, helicase, methyl transferase, and unlike structural/accessory protein-encoding genes, such as S, M, E, N proteins, that are located at the 3’ end which exhibits excessive variability(Figure 1).23,24Among non-structural proteins, most play a vital role in coronaviruses replication. Structural proteins, however, are crucial for virion assembly as well as for infection.25 The inhibition the activity of 3CLpro would block SARS-CoV-2 replication. The Xraystructures of this protease from SARS-CoV-2, such as PDB:6W63; 6M2N; 6M2Q; 6Y2E; 6Y84; 6YB7, are available on protein databank (Figure 1).
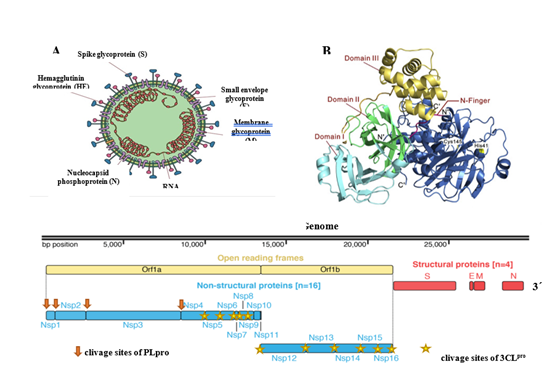
Figure 1 SARS-CoV-2 structure: SARS-CoV-2 capside showing major proteins and single-straded RNA (A), the model of 3CLpro a SARS-CoV-2 drug target (B) and (C) genome of SARS-CoV-2 and the large replicase polyprotein P1ab. The figure A, B and C were adaptaded from Moonchak V Projecto, references 29 and 47 respectively.
SARS-CoV-2 and SARS-CoV share a very high (96-98%)sequence identity for their 3CLpro and is located in a highly conserved region of coranaviruses genome.Therefore, it is a potentialtarget for anti-coronaviruses inhibitors screening and structure-based activity analyses and highthroughputstudies have identified potential inhibitors for SARS-CoV and MERS-CoV3CLpro.26 The structure and activity of SARS-CoV3CLpro has been unveiled and prompted the design of 3CLpro inhibitors as novelantivirals. SARS-CoV3CLpro features three domains: I3CLpro (comprising residues 8–101), II (comprising residues 102–184),and III (comprising residues 201–301). The active site regionspans domains I and II, which are β-barrel domains, while domainIII displays an α-helical organization. The cysteine residue 145(Cys145) of the active site behaves as the nucleophile, while thehistidine residue 41 (His41) functions as the general acid base.27
Drug development anti-SARS-CoV-2
The scientific communityis investigating new drugs against SARS-CoV-2 and have come up with three strategies for developing: the first one is assay existing broad-spectrum anti-virals, such as interferons, ribavirin, and cyclophilin that are used to treat coronavirus pneumonia. The great advantage of these treatments is the biochemistry, pharmacologic, pharmacokinetic, dosages and side effects are known and they have been approved for the treating of viral infections. However, they are broad-spectrum and cannot kill corona viruses in a targeted manner.28 The second strategy is to use existing molecular databases to screen for molecules that may have therapeutic effect on SARS-CoV-2. High-throughput screening makes this strategy possible, and new functions of many drug molecules can be found. The third strategy is directly based on the genomic information and pathological characteristics of different coronaviruses to develop new targeted drugs from scratch. Theoretically, the drugs found through these therapies would exhibit better anti-coronavirus effects.29
Many compounds have been assayed against SARS-CoV-2 3CLpro using combined structure-assisted drug design, virtual drug screening and high-throughput screening30 and some examples were discussed here. Pyridine-containingα-ketoamides, presented good pharmacokinetic properties in animal model and were detected at high concentrations in lung tissue and broncheo-alveolar lavagefluid within 4 –24 hours after subcutaneous injections, demonstrating important lung tropism. Besides sub-cutaneous administration, inhalation of nebulized resultedin high and long-lasting (24 h) levels in lung tissue, withoutany adverse effects, suggesting that these α-ketoamides are potential candidates in COVID-19 therapy.31
A mechanism-based synthetic peptidomimetic inhibitor of 3CLpro, a Michael acceptor inhibitor N3, was identified by computer-aided drug design that can inhibit the 3CLpros of SARS-CoVand MERS-CoV,32was shown to form a covalent and irreversible bond with SARS-CoV-2 3CLpro.33 Furthermote, the high-throughput screening approach for identifying inhibitors of SARS-CoV-2 3CLpro, ebselen, an organo selenium compound with anti-inflammatory, anti-oxidant and cytoprotective properties, was assayed with N3. They had expressive antiviraland host cell protection activities demonstrating their potential against SARS-CoV-2.33
The described inhibitors from different coronaviruses 3CLpros can be employed as anti-SARS-CoV-2, due to the high similarity of this protease among coronaviruses, as stated before. The majority of these inhibitors are bindcovalently active-site cysteine, including α-halocarbonyl, acrylamides, sulfonyl chlorides, aldehydes, isatines, or α-ketoheteraromates.34 However, they did not fit very well into the active site, because they are not peptide substrate of the protease. Interestingly, some compounds noncovalently binding to the active site of the 3CLpro seems to be a better strategy. A high-throughput screening identified pyrazolidinones, nitroanilides and α-aminoacylamidesas SARS-CoV3CLpro inhibitors with good antiviral inhibitory. But these molecules suffer from extensive metabolism and rapid clearance. Nevertheless, they are a promising starting point for further drug development.35
The anti-coronaviruses activity has been investigated in many plant-derived natural products and flavonoids, with diverse biological activities, have been shown to inhibit the 3CLpro.29Quercetin, a flavonoid compound, is widespread in fruits and vegetables, displays a great diversity of biological activities including anti-inflammatory, anti-oxidant, anti-viral, anti-allergic, anti-cancer as well as vasoprotective.36 Studies have found that quercet in exhibits antiviral properties against a variety of viruses, including Influenza A Virus, Hepatitis C Virus, Enterovirus, and SARS-CoV.37It has been confirmed that quercetin showed a good inhibitory effect on SARS-CoV3CLpro, about 82 %, expressed in Pichia pastoris. In addition, enzyme inhibition assays in vitro also showed that quercet in had inhibitory activity against SARS-CoV3CLpro.38 The 3CLpro sequence of SARS-CoV-2 is highly similar to that of SARS-CoV,28 thus, quercetin might have antiviral effects on SARS-CoV-2. This flavonoidwas docked to 3CLpro and was a good binder with energy of -5.6 kcal/mol. It also bind to Sprotein, ACE2, RNA-dependent RNA polymerase and PLpro indicating good potential against SARS-CoV-2.39 Quercetin has a wide range of sources with relatively low cost, so it is worth testing its efficacy against SARS-CoV-2 infection. Besides, many other plant derived compounds from Chinese herbal medicine and commom use such as andrographolide (labdane diterpenoid from Andrographis paniculata), glycyrrhizin (glycosilated saponin from Glycyrrhiza glabra), baicalin (glycosyloxyflavone obtained from Scutellariabaicalensis), luteolin (3′,4′,5,7-tetrahydroxyflavonecommon flavonoid), tanshione and derivates (abietane diterpenes isolated from Salviamiltiorrhiza), kampferol (natural flavonoid), hesperidin (flavanone glycoside found in & nbsp citrus fruits), and betulinic acid (naturally occurring pentacyclic triterpenoid) was also docked to 3CLproand demonstrated to be very good binders of this protease. However these compounds were not specific to this protease and interacted to others viral targets, thus, they can be considered excellent adjuvants in the treatment of COVID-19.29,37,39,40
The approved HIV protease inhibitors were previously repurposed for the treatment of the SARS outbreak in 2002 to 2004.41 So, they were hypothesized to inhibit the SARS-CoV-23CLpro. These drugs were docked inside the SARS-CoV-2 3CLpro. Importantly ritonavir, lopinavir, and darunavir had very good docking score of -8.878, -8.358 and -7.208, respectively. These three inhibitors are being tested against SARS-CoV-2.42 However, HIV protease is anaspartic protease and differs considerably from 3CLpro, but it also shares some common elements, such as a tetrahedral transition state and receptor pockets to recognize the amino acid side chains of the substrates.42,43
Peptidomimetic seems to be more potent inhibitors to 3CLpro. They have been used to treat many diseases like cancer, diabetes, autoimmune and viral and have high success rates in commercial development.44 These peptide inhibitors mimic a peptide bond and are better inhibitors than the others compounds. They bind to active site of SARS-CoV-2 3CLpro by multiple points in a combination of hydrophobic, hydrophilic and charged residues holding with hydrogen bonds. Peptide-like molecules provide a basic pharmacophore for the design of SARS-CoV-2 main protease inhibitors. The amide linkage backbone gives them the flexibility to fit confortably inside the binding site. Peptides may be the best alternative for small molecules for COVID-19 treatment, because they have great structural diversity, are easy to synthesize and are much less toxic when compared to synthetic and natural small molecules.45,46
A great diversity of compounds, synthetics or naturals, new or repurposed, have been extensively investigated against different targets of SARS-CoV-2 to develop drugs and pharmaceutical formulations in the shortest possible time to face the current pandemic. The 3CLproseems to be one of the best targets in coronaviruses, because the amino acid sequence of this protease is very similar to the 3CLpros from SARS and MERS, coronaviruses that infect humans and cause letal diseases. Besides, the 3CLpro inhibitors, specially the peptidomimetics, would be more specific to viral protease than the host enzymes, because 3CLpro-like proteaseare not commom in the human host. Thus, 3CLpro-baseddrugs for the treatment of COVID-19 would be expected to be more tolerable for humans due to the fewer sides effects. However, this questions will only be answer after biological and clinical studies.
None.
The author declare that there is no conflict of interest regarding the publication of this paper.
None.

©2020 Silva-López. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.