International Journal of
eISSN: 2573-2889


Research Article Volume 4 Issue 3
1Kazan Federal University, Russia
2Karunya Institute of Technology and Sciences, India
3VSB Engineering College, India
Correspondence: Rajasekara Ekambaram, VSB Engineering College, Karur–639111, Tamil Nadu, India, Tel +91 9524825876
Received: June 03, 2019 | Published: June 28, 2019
Citation: Rajasekaran I, Rajasekaran M, Velusamy K, et al. Drug protein interaction validates the internal cod formed due to cohesive force: test of bond length variation in amino acids involved. Int J Mol Biol Open Access. 2019;4(3):113-117. DOI: 10.15406/ijmboa.2019.04.00106
Background: Cross validation of internal COD obtainable from carbon rule of law that govern human body to function are crucial for future application is concerned. Verification leads to identification of all sick cells to cure to outperform better tomorrow. Cross validation of bond of all atoms is necessary for checking and arriving at conclusion that whether disease can be cured or not. Importantly rule of carbon based law interpret that incoming probe can be altering internal COD value obtainable from carbon value alone. Cross checking of these internal COD obtained from crystal data are to be the focus of this work. Verification can also be done from internal COD of protein in native form and alternative one obtained via complex formed using probe-protein interaction. Results are in agreement with cross validation of protein in two different states via internal COD obtained from crystal data of bound and unbound protein. Fantastic results are in agreement with changes of internal COD and bond of changes at that point. One of the mini tests of internal COD identification obtainable from sequence alone is demonstrated here in addition to original work. All are in accordance with rule of law obtainable from carbon value 0.3144. Results are promising that futuristic application that leads to disease free world of human nature.
Keywords: carbon domain, internal COD, bond of all atoms, drug protein, cohesive force, pentacarbon, protein research, bond order value, carbon value, fraction of carbon
Amino acids form a series of encoded message in dealing with atmospheric nature of biological conditions, otherwise call it as the plasma of human blood. When some foreign bodies enter into this system of biological nature, there is going to be chaos adequately dealing is needed for control of such action called drug therapy. One such an action is heat control of body temperature by paracetamol drug which alters biochemical processes in dealing them in which the application is specific in nature whereas other system cannot be interfered by drug or the other way around of taking away the drug without doing its job of temperature control. Specific in the sense that molecular level of interaction is focused here. Nonetheless the interference coming from drug can be altered in due course of action by any other molecule which regulate body metabolism. Overall performance can be monitored for existence of foreign input in due course of action during alteration, otherwise call it as evolution of human kind in existence for co-editing arranged reaction to occur. In a simple term arranged reactions are to be happening according to the nature of law that are in place from time to time. Fantastic of those reactions are said to be forces of interaction. Notwithstanding all those happening in human nature specific needed to be addressed. Here is force come into existence which is in due course of carbon rule based fraction of 0.3144 value.1 Co-existence can be adequately controlled and monitored for disease control. Here is again cohesive force arising out of carbon rule of law dealing this phenomenon of nature of carbon value.2,3 Very well understood that carbon can be the key role of player of all living organisms and core needs to be addressed from rule of law coming from carbon alone which is taken up here for further validation from bond of all atoms. Accordance with rule of law that carbon can be arranged according to principle of maintenance of carbon value which is close to 0.3144 everywhere in the system of dealing. Adequacy is the principle of carbon based law that addresses all macromolecule to interact and deliver things.4 Nonetheless adequacy leads to stability 5–7 and address all phenomena of binding followed by biochemical processes.8 Adequacy can be monitored via carbon value based on domain formation and some time atom’s bond lengths. Arrangements are that according to nature of law that adequacy is maintained all across system under control. When the probe enters which alters the due course of action according to the rule of law again. Arrangements are that any other probe may not be able to interact with internal matter of protein system. Overall functioning is fine with adequacy principle. Whereas one such function deviate means the entire system of action falters accordingly. Even though adequacy principle govern them all arrangements are not provided for similar one to perform opposite reaction9,10 which are taken up here as case study. Study reveals that adequacy is dealt with one portion of drug whereas other portion can be participated into new beginning of uncultured reactions which are evidence of several viral based interactions in the human system of evolution which again needed to be addressed and asked for validation. Cross validation of such involvement can be arranged according to the rule of law that arises as part of our study. One such a biochemical reactions ruled by small molecules is demonstrated here as case study and alteration can be discussed in terms are futuristic applications. Main thing is to validate the internal COD obtainable from carbon rule of law and capture the phenomena of alteration at protein molecule based on probe binding. One such a system is arrived at to demonstrate all atom level changes are due to internal COD and apply for futuristic application of drug delivery of disease control of arranged molecular interactions.
Input
The protein structure files are retrieved from protein data bank of RCSB. The details of retrieved one are given by the following. The protein in native form (1kas) and complex formed (1b3n) are detailed at NCBI and EBI web sites where one can get all other information regarding this protein of interest. Overall one can obtain the necessary information about morphological parameters. Whereas we are in need of the internal parameter which is of focus here in this calculation in our laboratory using simple in and out parameter obtained based on carbon value calculations. Otherwise the protein structure input parameters are incomplete due to non availability of crystal information derived from X-ray here where in one need to do away with crystalisable protein fact of all atoms. Where one of the amino acids missing at the beginning was ignored in our calculations. Secondly the atom at the bottom of the coordinate file contains no OXT data, deleting in both the cases for uniformity. Sulphur atom at CYS163 is ignored during addition of hydrogen atom by online software tool molprobity. Otherwise over all it is fantastic to use for calculation of cohesiveness via internal COD (Carbon Optimized Domain) in the protein in native form and complex formed.
Internal cod for diameter of 16å
Internal COD obtained via CARd program are reported earlier in our other papers. Whereas here our purpose was to see the obtained ICOD value variation due to complex formation and validate via bond of all atoms. CARd16 program reads our coordinate file, convert it into array form for further processing, ignoring all other parameters for calculation. It is standalone program developed by PERL set of instruction for coding and validation. Outsmart other calculation by reading coordinate into array without any hinges for calculation dynamically. Arranged are the coordinates and obtained was a set instruction to be followed for mutation or other to follow which is automatically can be done over a computer. Arrangements are to give set of instruction to be followed. To be precise the internal COD are given at amino acid positions where by obtained values are validated using bond length parameters including Cα-C bond of back bone atoms obtained via crystal data where in one would require only static value for dynamic prediction. Otherwise call it as parameter of nonsense data to go with static one. Overall out perform well so far in prediction verifiable by our data. Number 16 may be important here for prediction of internal COD for validating bond of all atoms. All that happens at diameter of 16Å where in adequacy principle is followed for satisfying carbon value for COD formation. Rule of law says that adequacy is met with 16Å diameter for satisfaction otherwise call it as non-COD portion. When in need of the outer parameter may be helping this achievement of adequacy carbon. Formation of COD everywhere is important for protein to follow adequately in water system of biological nature. Placed in somewhere else resulting denaturation of protein fold for adequacy concerned. Organic solvents treat this adequacy principle differently overall here our performance is concerned. When in need CARd3D can be modified for treating these situations to deal with other solvents and validate parameters of interest from physical properties obtained via calculations or from experiments. Necessary changes needed to be obtained via PERL instruction for changes from time to time. Automation is need of this hour for all those to happen automatically obtained every moment of instruction to be followed where in need based on instruction obtained via rule of law governing everything altogether proper changes are to accommodate authentically otherwise neglect it for change instrumentally adequacy of carbon principle of rule of law obtained via COD data obtainable from static values. Whereas one might want to go for early prediction of obtained data single handedly wherein data alone is sufficient for calculation.
Bond length cross validation
Programs based on static data are over written to snap taken for bond of all atoms which is of validation for internal COD obtained differently by calculation of all atoms to be included during calculation. Static data provides details on dynamic nature of bond length variation with respect to internal COD obtained via 16Å diameter calculation where in internal data influence of 16Å diameter long is obtained one. Specific that internals are reflected in bond of all atoms. Directly and indirectly influences bonds of all atoms provided greater is influence of cohesive one formed via internal COD obtainable from adequacy of carbon value where in requirements are the scale parameter called 0.3144 value of carbon fraction verifiable from all other calculation obtained from data of static in nature single handedly overall for biology is concerned. Man of earth can be better tomorrow provided the validation of these parameters are adequately given importance all over the system of functioning body of human nature. The obtained bond lengths are figured using excel point, including static values at COD or non-COD regions which is of great influence from cohesive point of view that carbon causing this changes adequately at each position of interest.
Internal COD assessment based on carbon values of pentamer amino acids
Assessing internal COD is one the challenging factors for all assignment to be departed. Accordingly one might want to assess from carbon alone which is cheapest in computational point of view where as internal COD of all atoms takes about few seconds averaging all over again according to our program written using PERL standard whereas one might be able translate into another program where calculation to be in milli seconds. No matter what it is demonstrations are here to stay in the event that anybody might want to work with existing one suggesting any improvement over the issue of development costing too much to consume additionally. Otherwise it is in agreement with existing one to go on further.
Internal COD by diameter of 16
As can be seem from Figure 1 that the internal COD values are plotted against amino acids involved in it. First of all the internal COD value indicate whether there is domain or not. Zero value means non domain region. Rest of the portions whether it is small or larger value, it is a domain region. One can go on explaining active and domain regions based on internal value calculated from CARd program described in reference.1,4 But here what happens to these domain or active regions upon complex formation with molecular probe which may be metabolites, drugs or any other small molecules. Active regions are supposedly changed upon binding of incoming probe pattern appropriately. One can expect a non internal COD becomes COD during complex formation which is happening here as the probe, cerulenin interact with protein synthase. Details can be as described here below.
Figure 1 showing results for internal COD values of synthase in both bound and unbound state. As can be seen in blue colour the native protein demonstrates the internal COD values along the sequence of amino acids in the protein. Here again the internal COD values that are non zero are domain portions. But the non internal COD portions seen at four places, 38-46, 106-111, 294-298 and 377-383 for example are non obedient to COD formation. They are likely change during external molecules is in force which we observe here as can be seen in dark brown colored line obtained for probe bound state of protein. Where external forces cohesively changes the internal domain values, mere alteration in atomic coordinates locally and so forth. Binding here in one location of protein, changes everywhere in the system including those non domain portions and all of the other sites as well which may or may not yield another active portion which may be essential for further action in the system of reaction biologically. Meanwhile there may be non domain portions which are essentially included in the complex formation where incoming molecule can be fit in to release probe molecules when there is need of such action which eventually leading to synthesis of molecules retaining the original molecules without any changes.
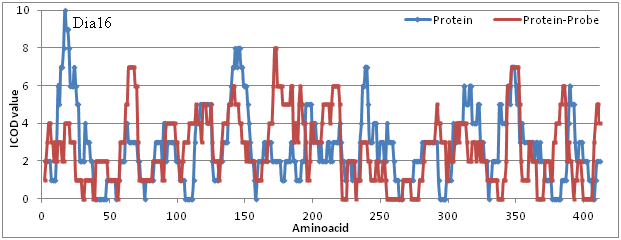
Figure 1 ICOD for native and complex formed protein. Noting that where ever the blue line of zero ICOD value then there is non-COD portion demanding probe to bind to satisfy them all. Note also that the dark brown line satisfy them all making it to COD of all atoms where by ICOD dominates all over dealing stability of protein. Strong bond formation adequately contracting for all to happen again meaning that protein fold better over the counter part. Protein folds because of this ICOD value in it due to cohesiveness accordingly both in terms of local and global changes. Better known as folding force ICOD value to fold protein in the native form and relative to one that is complex formed with probe.
Obviously, there are domains which will no way change in non domain that can off late be called as pattern of conserved regions which can be collected later on mutational studies leading to protein stabilization followed by specific function without interfering the other system of molecular association which is essential for long live biology of ego system where internal wastage can be prohibited for long time to come where in probe binding be specific accordance to the law of requirement adequately at affordable cost of living in the universe of human system of workable condition in the atmospheric air. Nevertheless according to rule of law that governs protein fold may be of great interest in the near future for manipulating human elevation to humanoid into the universal kingdom for internal matter is concerned. Very good observation is that man can be verified by potential signature on internal matter that can allow man survive better over the current issue of disease suffering and other age related matter of salvation in the universe of human kind that can outperform everything without hindrance at affordable cost and in due course of action universally.
Bond length validation
Bond of all atoms can be compared to validate the existence of internal COD in the protein of interest. There must be a care taken adequately interpret the results of all atoms that may be involved in COD and/or non COD region of interest where bio-molecules interact to bring about biochemical reactions. Adequacy is met with internal COD during cohesive formation internally. All that happens at a time adequately without intervening of other forces of interaction adequately dealing the phenomena of biophysical properties. Nevertheless carbon based COD can be of great use in explaining tall he details of molecular association or fold themselves to attract incoming molecule of potential application. Whereas the other forces of interaction can be negligibly strong enough to perform automated validation of protein bond of all atoms. CARd program dealing the COD of all atoms can be better validated with the help of bond association between any atoms where one would focus. Currently we are focusing on back bone Cα-C bond where one expects all adequacy principle accumulate for changes. Moreover carbon-carbon single bond can be validated for expansion or compression due to the internal COD formation. Obviously other forces can be included there to verify internal changes. One would want to do serious calculation in performing these cross check. Whereas carbon domain is a measure of bond length validation in which direct evidence is put forth here. As a measure of COD formation one can compute the bond length at a point where COD of all atoms need to be considered. Never mind taking all bonds into consideration for measure of COD formation. However bonds are responding differently with given surrounding. Alpha carbon of the back bone chain may of great use in dealing such situation. However care must be taken here to consider back bone carbon-nitrogen bond which is of opposite response with respect to COD is considered meaning that the partial double bond expand on COD formation over the issue of non COD one. This is because of the delocalization of double bond electrons into COD of all atom. Whereas very near to double bond character of peptide nature, there is another carbon-nitrogen bond which is also along the back bone chain that never altered as in peptide bond meaning that double tend to go single bond while single bond pact with COD of all atoms. Surprise to see these results including in aromatic in nature where delocalization is possible by COD formation which can be a measure of all atom involved in it. Whereas measurement means bond of all atoms but differ in interpretation of results of carbon domain which is of great use to us. Nonetheless carbon-carbon single bond alone can be of measure for bond of all atoms calculation which is done here.
The Figure 2 shows bond length variation among Cα-C bond of individual amino acids in protein crystal. Where the blue line indicates the native form of protein and dark brown indicates the changes with probe association. Meaning that bond association can be a measurable quantity of internal COD formation. Locally there are bond length changes with electro negativity atmosphere. However our COD formation can be felt at 16Å or so. There is remarkable change with bond length variation at this internal COD of diameter of 16Å which is matching very well with internal COD observed. It gives impression that internal COD can be directly measured with bond length calculations.
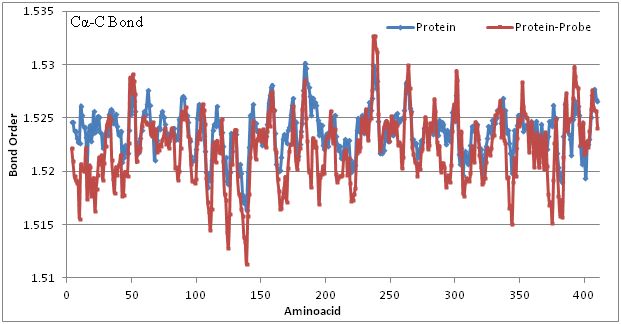
Figure 2 Graph showing bond length variation along the protein sequence that is bound to follow ICOD value. Blue colour indicates the Cα-C bond order for native protein whereas dark brown shown for complex formed by protein and probe. Almost all places there is decrease in bond order meaning that the complex is more into internal COD than that of native form. Otherwise ICOD favors bond length shortened due to cohesive force taking away the possible electrons of COD formation. ICOD can be a measure of cohesive force involved in it. Blue follows the bond length uniform everywhere but dark brown are varying.
Bond length decreases everywhere due to the probe formation complex with the protein describing it as COD all over the molecular protein. However detailed analysis describes that a zero internal code portions are having higher bond length termed it as active region and give counter effect of bond length variation in the adjoining neighbors during complex formation where higher internal COD values must be decreasing the bond order. Specific to the active site of zero internal COD value (called it as non COD portion) where the incoming probe turns it a internal COD then there is decrease in bond length. It can be observed at 38-46, 106-111, 294-298 and 377-383. Similarly, when COD value increases or decreases at some point where there is no active stretches then the bond length decreases or increases accordingly. Diameter 16Å is so significant at this point of observation here. Once again when changes take place at active regions, the counter goes meaning that the neighboring amino acids are cope up with new developments adjusting bonds for non COD.
There are four different situations terming COD to non-COD in general and at adjoining neighbors of active form and non-COD to COD in general and at adjoining neighbors of active form. Out of these always non-COD to COD decreases the length and vice versa. Adjoining neighbors give counter of these that non-COD to COD increases the length and vice versa. Internal COD including diameter of 16 and bond lengths go hand in hand as evidence from this protein and protein-probe complex comparison.
During complex formation COD turns non-COD forming active region or the vice versa may not be point of discussion with length variation since some time increases or decreases and adjoining neighbors are important here as it give counter effect. The adjoining one increases bond length due to COD formation and vice versa that are opposite of conventional one for general one. Upon binding of probe the increase in COD value decreases the bond length and vice versa but COD into non-COD also decrease in value which is not increasing bond length but counter effect. This is because of non-COD of domain formation with neighboring amino acids. The Non-COD active stretches at 38-46, 106-111, 294-298 and 377-383 turn COD upon binding expectedly. Bond lengths are decreased adequately. Internal COD portions at 255-266, 273-278 and 400-406 turn non-COD during complex formation. No change or slight increase in bond length.
Short internal COD at 30, 224, 232, 338, 368 and 393 turn non-COD upon binding leaving lower bond length to further lower and/or higher one to further higher. Middle ones remain unchanged. All other internal COD respond to bond length variation accordingly that the higher internal COD at 239 and 316 turn out to be lower internal COD but remain COD and bond length increased accordingly. Same way lower internal COD at 177, 188 and 213 turn higher internal COD and the bond length decreased accordingly. Whereas the increase in internal COD at 66 and 386 are adjacent to active stretches around 40 and 378 respectively. So is the counter effect that is increased bond length. Accordingly the higher internal COD at 31 and 390 turn out to be lower internal COD and adjacent once again to active stretches around 40 and 378 respectively. This again counter effect and decreased bond length.
By definition, active sites are one that is non-COD of all amino acids involved in which undergo changes upon mutation or external binding. Non-COD regions are responsible for binding and carry out with activities of biochemical process.
Analysis by pentamer amino acid’s carbon values
One of the recent developments here in our laboratory that averaging carbon value of pentamer can aid in understanding active non-COD and carbon domain regions. We have developed a program called PentaCarbon for averaging carbon values and analyse further on internal COD values. Relationships between these two are observed. Fact that carbon rich portions are assumed to be active form and vice versa. Very good in agreement at some point whereas the carbonless portion can also be forming active sites which is demonstrated here at 106-111. In the unlikely that monomer value of carbon details everything but pentamer value, sometime dimer also in pray for calculation. Overall dimer to pentamer can be of great use to us in assessing internal COD values whereas pentamer is demonstrated here along with internal COD of protein interest and bond length values.
One of the interesting phenomena of binding studies reveal that protein fold and bind according to the nature of internal COD available in the protein system of interest, unlike the one studied recently in our Kazan Federal University, Russia11,12 that the peptide of four or five amino acid interact cohesively to form hydrophobic nature interaction. Withstanding to those modifications that protein itself needed to be focused for any drug development. Experimental evidence may not be good enough to deal with this large set. Dealing computer based method for arriving at conclusion that phenomena of binding may be help from development point of view. Similarly one the fundamental fact that protein associates and/or dissociates based on the principle that carbon role in internal COD followed by cohesion. Additional information may not be required to address these phenomena of protein fold and binding. One such a study in the recent years for validation of target protein from fundamental concept of drug-protein interaction is arrived at by using other forces of interaction which may yielding any fruitful results. With this carbon force formed by lining amino acids fundamentally is so strong for future full of application and narrowing down the disease pathway of molecular interaction.
Overall performance of carbon value obtainable via internal COD is validated. Single handedly works very well with bond of all atoms verifiable from carbon rule of law governed by internal COD obtained from carbon fraction 0.3144. Otherwise call it as great use of bond length variation with respect to internal COD and cross validate the internal COD in deserving places and/or not in deserving places from pentamer value of carbon fraction. Verifications are excellent in outperforming adequacy principle of carbon rule of law obtained from carbon alone here where in biology is concerned. Human can excel in all other performance provided cross validation is obtained all over the system of functioning body. Hopes are there to see this happening in the near future of fantastic applications.
None.
The author declares there is no conflicts of interest.

©2019 Rajasekaran, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.