International Journal of
eISSN: 2573-2889


Research Note Volume 3 Issue 5
Department of Chemistry, V.S.B. Engineering College, India
Correspondence: Rajasekaran Ekambaram, Department of Chemistry, V.S.B. Engineering College, Karur-639111, India, Tel +91 9524825876
Received: September 20, 2018 | Published: October 25, 2018
Citation: Ekambaram R. Domains based in carbon dictate here the possible arrangement of all chemistry for biology. Int J Mol Biol Open Access. 2018;3(5):240-243. DOI: 10.15406/ijmboa.2018.03.00083
In recent time, I have reported that the existence of carbon value in carbon domain is determining to be the factor of all stable regions in a given protein here. Again conclude that the Globular Amphipathic Domain (GAD) based on carbon value is observed to be the fact of maintenance and responsible for stability of the protein and activity. CARd program explains all this phenomenon of protein to fold. Average value of all possible outerlengths is looked out here. Analysis of some enzyme proteins, including super oxide dismutase and ribulose-phosphate 3-epimerase are looked in details here up again. Normally a protein folds because of this GAD. Observed that GAD dominates of all forces of interactions that has been discussed so far. Hope to find solution for folding of protein to occur all time.
Keywords: carbon value, icod, reduced-carbon, carbon-high, carbon
Protein sequences are the primary sources of all related information buried inside a given protein1. At the same time persistently conserved positions in structurally similar and sequence dissimilar proteins preserve protein fold and function2 adjusting both in order/disordered one.3 Evolution dominates the available buried information for function of protein.4 All available functional units are few residue lengths conserved of all dominant force.5 Changing these residues might be hydropathy focusing point of view that this conserved dominant force of attraction present is broken. All neutral residues are likely to be critically important for maintaining optimal functional variation of protein function of all time.6 Hydrophilic of these functionally important portions might be the critical sources of disease all time in. Hydrophobic interaction is seemingly important for protein to fold among themselves7 and in diseased proteins. The importance of slow motions for protein functional loops8 related to hydrophobic interaction is needed to be discussed at this hour. Functional unit coming from meta sources might be the critically diseased if it serve for meta protein of all time sources. According to meta sources of all time high visible interactive media focus of all alone sequence information, the emphasis is given for the diseased one. Enzyme for example is focused more in the sense that the chemistry of catalytic action being the focus point of all time. Given the situation hydropathy focusing point of view, a rational view point for catalytic site of all time focus is needed at this juncture. Sequence being the source of all time hydropathy focusing point of view, ration has to be devised for that point of view from active amino acid all along the sequence of interest. Good that there is no direct measure of hydropathy focusing point, but indirect one is devised for that. All along the measurement, focus is being given for end that is being focal point of view in the sense that views from the hydropathy point of view measured. Calibration is being focal point on that device of measure which is measured directly on mechanism of focus. Sequence for example, all focused on measurement of binding capability of individual amino acids that are focal point of view of the individual management system that are individually measured. Mutation for example focused more than that of direct measurement of all time focused individual alteration focused in amino acid of interest. Models are central focus of all time binding capabilities of individually carved one that is being focused on binding of individual amino acid of interest that is taken up here for study as central focal theme of interest of all time focused individually at amino acid level of interest of all time here.
Super Oxide Dismutase (SOD) is taken up here as central focus of individual study of interest. Here in after called reference protein, point of view for that all available information is suitably noted on the central theme of study. Focus is emphasized on novel development of parameter for fixing all available sources of information that are being currently used up here for focusing the new developmental activity of interest. Emphasis also been given on secondary development of additional point of view that any available sources that can be deployed additionally. Additionally where there is no availability of conflict of interest, the ribulose-phosphate 3-epimerase is taken up here for further understanding of further information of all time here to stay.
Focus has been emphasized more often on the deployment of additional information available from various sources of interest that are being collected individually on the basis that individual emphasis is given attention of all time. Also focused on deployment of newly developed technique called CARd9 that’s been there for a while on the online tool development. Emphasis is given on fixing the program on focal point of view that it fixes up all parameter of interest in biology by narrating the chemistry of all time. Emphasis also been focused on deployment of available sources of interest for new development of interest. Additionally anything new on productivity of additional information can be focused on the basis that it suitably noted in all respect of all additional point of view of the protein. Primarily it focuses on sequence analysis of protein system that can demonstrate all other sources of information available from point of view that it can work out very well on emphasis of newly developed productivity line of interest at all time of interest. Sources of newly developed productivity line are available for other sources of interest for other applications. Note has been given on all available interest of productivity. One can deploy field of study of interest on emphasis of interest of all time. Also available are deployment of all time deployed parameter that can be fixed up at all level of interest. Please make a note of it at level of interest from the point that it is noted everywhere for personal use of interest of all time of all level. Emphasis can also be given on local development of own product of interest for all applications.
Data
The sequence of structure of super oxide dismutase (here in after it is called reference protein) retrieved from PDB.10 It is a NMR structure containing 153 amino acid in length (PDB:1DSW). Another sequence to capture the essence of structural information is selected from Swissprot/UniProtKB11 where there is no information about its structure. It is being the ribulose-phosphate 3-epimerase. Reference being Q9ZTP5 and crystal elucidation is impossible for this.
CARd analysis
CARd analysis on the protein sequences are carried on using well known program called CARd. Details can be found at Pubmed of NIH.12 The flow diagram (Figure 1) outlines how the distribution of inner lengths based on carbon fraction is counted in an outer length. The pink coloured sequence protein sequence which is converted into atomic sequence (shown in multiple colours (red+blue+pink). The red portion is an inner length. The blue portion is outer length which includes the red portion as well. The entire atomic sequence is given in pink colour which includes both blue and red. Here the outer and inner lengths are taken as 100 and 35 atoms. There are 65 (100-35) inner lengths for statistics. These 65 inner lengths are grouped based on carbon fraction. The C11 has (11/35) 0.31 carbon fraction. The program named CARd has been written in PERL programming language to undertake this entire task. The program reads the protein sequence, converts into atomic sequence, takes a length (anything from 45 to 700 atoms) of sequence, divides into small lengths (of 33 to 350 atoms) of equal sizes, finds fraction of carbon atoms in all these small lengths and counts number of small lengths that contain a defined fraction of carbon. There are small lengths with 0.250/0.450 carbon but maximum at around 0.314. A distribution of range of small lengths based on carbon fraction appears like a normal distribution curve. This distribution curve is obtained for all possible outer length. The outer lengths can be any length within the range of length between 45 and 700 corresponds to 3 and 45 amino acids. Any outer length chosen between 100 and 250 atoms is sufficient for most of the observations. Here in modified version of CARd, it is selected as 62, 78, 109, 125,140, 155, 171, 202, 218, 233, 249, 264, 342 and 700 in our calculations. The inner length has been fixed at 35 atoms and can be anything between 33 and 350 which are not exceeding half of the outer length. Inner length of 35 atoms is chosen here in all our calculations as it is the smallest unit with 11 carbons which can produce fraction of 0.314 (close to standard of 0.3145). The outer length is moved with step value of selected atoms. Normally it can be half of the outer length. Here it is chosen as 7 atoms to capture carbon values at all amino acid location. A carbon distribution profile is obtained for all demonstrated outer lengths. So the carbon value obtained in terms of statistical valuation, namely mean, median, mode and standard deviation is graphed using XY plot. In fact it is only the median value taken as carbon faction at amino acid positions. Graph gives the idea of location of Globular Amphipathic Domain (GAD), carbon-high hydrophobic regions and reduced-carbon hydrophilic regions. If the values are close to 0.3145, then it is GAD, otherwise either carbon-high or reduced carbon portions depending on the hydrophobic values.
Three dimensional CARd analysis
CARd analysis using protein sequence alone gives that the GAD regions. The same thing can be measured using PDB structure of the given protein. Details again given online as CARd3D13 program in Pubmed of NIH. Slight modifications are carried out to capture the Internal Carbon Optimised Domain (ICOD), which is being equivalent to GAD. Modification includes an average values for variety of diameters for up to 45Å. Here it is taken for short range (7-16Å) only that captures the phenomenon of ICOD. Particularly it is evaluated up to 18Å for averaging. Graph can be drawn to visualize ICOD portion and so on. ICOD is one where participation of amino acid residues is dominant whereas it is low in non-ICOD portion that is being carbon-high hydrophobic portion usually.
Bond of all analysis
Bond length of all possible bonds are calculated and averaged for comparison. The reference protein is analysed with a term that back bone bonds are common to all residues which are adequately responding to the adduct forming ICOD regions. The bond length of all back bone bonds (such as N-CA, CA-C, C-N, N-H and C=O) are measured and the standard deviations are computed using standard bond length value considered in normal peptides. It is computed as by dividing the measured value by fact of 1. A PERL program which can accommodate all this phenomenon of capturing standard deviation of bond length is written, named as BondAll.pl and captured all bond length variations. In fact the double bond character of peptide bond is hoped to increase by fact of 1 in ICOD region. The other single bond character is hoped to be reduced further by a fact of 1.
The 153 amino acid long reference protein is subjected to GAD valuation. The calculated average median carbon values are plotted against residues (Figure 2). There is a long GAD region from 70 to 112 and after that. Similarly 1-14, 21-28, 32-39, 115-125 are GAD regions present in reference protein. There are carbon-high regions (for example 43-48) and a reduced-carbon region, 130-153. The GAD regions are hoped to be compact in nature, due to which the bond lengths are reduced. Fortunately, the NMR structure of this reference protein is available which has been checked for existence of carbon optimized domain (Figure 3). The result again confirms the presence of amphipathic domains. Infact it is internal carbon optimized domain (ICOD) based on structure that dominates the presence of amphipathic domains in sequence. Once again the variation of bond lengths confirms the presence of GAD, got obtained from sequence. These results based on structure will be put forth sometime again in detail. (Figure 4)
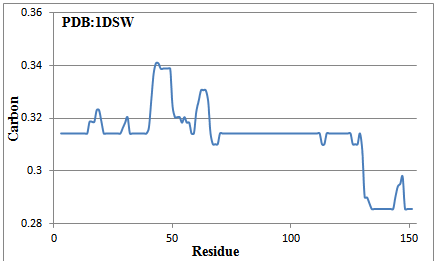
Figure 2 The median carbon calculated by using sequence information alone for reference protein. Note that there is GAD region at 70-112 and beyond that. Residues 15-20, 29-31 and 43-50 are carbon-high regions while 130-153 is in reduced-carbon region. The active region (62-64) is the carbon-high portion.

Figure 3 Participation of amino acid residues in icod for reference protein computed using pdb structure. Note that there is considerable amount of participation in ICOD at 70-112 and beyond that. Also note that there is reduction of participation in carbon-high regions at 15-20, 30-34 and 43-50. The carbon-high active region (62-64) is also less participation in ICOD.
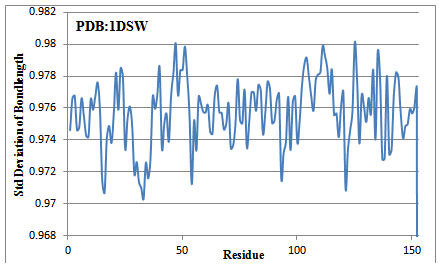
Figure 4 Average standard deviation of bond lengths of 5 back bone bonds(N-CA, CA-C, C-N, N-H and C=O) at each amino acid posistion. Note that the ICOD participating residues (70-112) have narrowed down its deviation considerably from standared of 1 and beyond variation occur that ICOD varying beyond residue number 112. Also note that carbon-high region 43-50 is showing less deviation than the other two hydrophobic regions 15-20 and 30-34. The carbon-high active region (62-64) is also in par with ICOD regions meaning that participating in ICOD in higher dias. The hydrophilic stretch 130-153 possess a variable standard deviation among the amino acid involved in. It is not a adduct unlike ICOD region but ICOD forming in.
A 274 amino acid long ribulose-phosphate 3-epimerase protein is subjected to GAD evaluation of protein structure. The average carbon value is plotted (Figure 5) against residue. There are several ICOD stretches including 46-58, 62-73, 98-106, 129-139, 160-170, 181-189, 212-217, 234-247 and 267-272. The binding sites are observed to be the character of hydrophobic in nature. The reported (refer UniProtKB: Q9ZTP5 for detail) substrate binding sites (192-195 and 247-248) are hydrophobic regions that are being adjacent to ICOD portion. Similarly, 56, 114 and 227 are reported as binding site. Except 56, rests are in hydrophobic regions. The reported conflict regions (211-213 and 267-268) are due to the presence of adjacent reduced-carbon regions. It is observed that the transit region (1-39) reported as in Swissprot are in hydrophilic character.
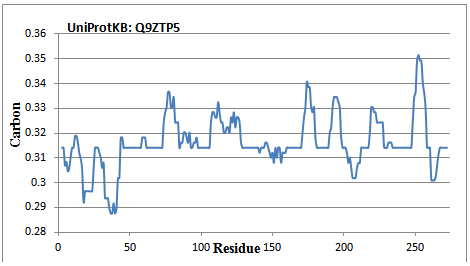
Figure 5 GAD computation which is being equivalent to ICOD in 3D structure for ribulose-phosphate 3-epimerase (taken from UniProtKB of ID:Q9ZTP5). There is a long portion (1-42) of reduced-carbon region. Reports being that the substrate binding sites (192-195 and 247-248) are carbon-high in nature that is being adjacent to ICOD region.
The ICOD forming bond length of selected back bone atoms are observed to be reduced in nature of all time. The internal amino acids involved in carbon optimized domain (ICOD) form a compact structure (adduct like behavior). The atoms are facing each other in such ways that meet the optimum value of carbon all over structure. The arrangement is to share electrons among themselves. The bond lengths reduced due to this in the structure. It’s like a Fermionic condensate, behaves like an n-type semi conductor, either conductor or nonconductor, but given the biological condition it allows electrons to go through and following the Fermi paradox. The adduct structure formed by this is behaving like as if aromatic compound that can be water repellent. This can oppose alteration by any other interacting atoms in biological condition. This fact needs to taken into account of new developments that validate macromolecular structure and function. Web based structure validation program that provides broad-spectrum of solidly based evaluation of quality of both global and local structure of proteins and nucleic acids14 are being considered here.
A adduct structure of ICOD comprise of amino acids having the above character in varying length. They are made from carbon and polar atoms of those amino acids organised in some facts of form. ICOD parts are small and may be complex and delicate geometry but others are large in sizes that are in the form of ICOD blocks. There are varieties of ICOD forming compositions evolved which have been adapted to a broad range of stability in proteins. Where ever it occurs in proteins, they are subjected to water repellent and conductivity. That is the linings of amino acids in ICOD build to water shield and electrical conductance. The ICOD stretch that has a high ICOD command is enough to swim while the protein is in function. All amino acids used in the forming of ICOD are important for water shield in biological conditions. Based on the application, ICODs must resist molecule of interaction, oppose chemical reaction, catalytic action and similar one. As variety of composition of ICODs instruct a variety of performance and properties, many ICODs have been evolved for different stability purposes.
Large varieties of ICOD are available. Only small ones are attracted with other interacting molecules. Larger ICODs are not important. Small five amino acids long ICOD will do for most of the local stability. Lightest and small amino acid series for ICODs are available. One can go for redevelopment of protein with higher stability and activity using this.
CARd program is extended here to demonstrate the existence of ICOD domains present in protein 3D structure. The programme is capable of capturing the essence of all possible stretches that include hydrophobic, hydrophilic, ICOD, binding, active, non-ICOD forming and amphipathic domains of all time. Successful completion of ICOD value determination is seemingly important from development point of activity for next generation to come. Observed that amphipathic domains are part of protein evolution that occur all around the structure and hydrophobic part that capture foreign bodies for interaction of all time.
None.
Author declares that there is no conflict of interest.

©2018 Ekambaram. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.