International Journal of
eISSN: 2573-2889


Research Article Volume 4 Issue 5
1Department of chemistry, VSB Engineering College, India
2Karunya Institute of Technology and Sciences, India
Correspondence: Rajasekaran Ekambaram, Department of chemistry, VSB Engineering College, Karur – 639111, Tamil Nadu, India, Tel +91 9524825876
Received: September 18, 2019 | Published: October 11, 2019
Citation: Ekambaram R, Rajasekaran I, Rajasekaran M. Domain formation in regions of protein probe interaction. Int J Mol Biol Open Access. 2019;4(5):167-169. DOI: 10.15406/ijmboa.2019.04.00114
One of the interesting focus points of research in protein science is the active site analysis and probe fixing. Overall one can go on searching this interesting phenomenon of probe fixing but without proper explanation on how and why this happens at some site neglecting other places. One of the probe fixing methods could be come along the way of newly discovered carbon role in protein stability that is taken here as starting point for due course of action. Carbon role in protein domain formation and active portion are interestingly yielded good result. Analysis includes ICOD of protein in native and domain state of active portion in complex protein. ICOD analysis reveals that carbon does play an interesting role in probe of action. Alternative method to detect active portion and probe development can be better exploited from this carbon domain of protein for life science to excel.
Keywords: carbon domain, ICOD, drug protein, protein interaction, carbon value, active site
Action of probe binding to wild proteins is available now to explore the possibility of extraction and alteration at active region for effective binding.1–3 One of the forces of attraction is coming from carbon alone. When carbon probe binds to active region, alteration taking place in the vicinity of probe binding and place of alteration in ICOD details. According to nature of law carbon must be adequately distributed evenly with value of 0.3144 4–6 everywhere in the protein. Probe can bind only when there is deficiency of carbon value according to rule of law. When there is inadequate carbon point of distribution where alteration can be performed by probe binding at the adjacent site effectively.
Accordingly initiatives are done to probe the role of carbon in dealing with other situations like, pattern recognition,7,8 disorders in protein,9,10 disease causing11,12 and mutational role11–15 when other forces fail to capture the essence of it. When in need one can probe those devisable mechanisms for possible arrangements to continue with carbon value based method of alternative design and action actually taken here. Carbon value is predominantly important for futuristic application of action of biological compound according to the need based system development. One can carefully analyse the action of adjacent amino acids which got altered during probe binding. Accordingly analysis can be put forth for futuristic application of adjacent amino acid of interest. Superposing of one on another might be yielding of interest in changes is concerned. If it happens at the site where probe can bind, well it is acceptable. But always unacceptable changes observed. If at all new mechanism which will define these observations, well one might think of carbon here which are defined here in this study.
The Protein (4JX3) and complex (2BZJ) structures are taken from protein data bank. It has 326 amino acids. All of them are not available with crystal structure. Some are missing even though sufficient amino acids represented available for analysis of active region. That is to say the first 32 amino acids and last few including 307–326 are missing element of amino acids. Given the structure, ICOD program can be utilized effectively to locate the carbon domain followed by active regions. The arrangements for ICOD analysis are carried out using CARd–3D program6 which again modified to meet out internal COD calculations. Options are wide open for various probe finding to drug discovery. According to nature of law of carbon value, one can extrapolate this decisive method of probe–protein interaction to any extent for biological applications but limited to crystal availability. Otherwise call it as application of aggressive analysis of protein and development of active regions. When in need animalia kingdom can be probed for better recognition and action of probe particle. Applications are successful for protein engineering but limited to other macromolecules which again give signal for post translational effect. Extension can be of useful for protein analysis and effective functioning all along the probe binding. Details are later in adequate timing of research will be put forth according to availability of other sources of input. Needless to say that carbon is going to roll out big impact in the society of research profile where in probe analysis will take over the other side of research. Fundamental shaking of probe–protein interaction is there to look into new courses to begin with. ICOD values are found out at dia 15Å, even though maximum happens at 16Å. Protein and the complex are analysed using our home made program written in Perl programming. Everywhere there is domain regions except active region and are discussed in the results section. Infact a feature articles discussing trends in targeting proteins with metal complexes are the starting point here. Active portion finding is the main focus of this work.
Carbon analysis profile for the individual protein and its corresponding protein ligand bound structures are analysed as shown in Figure 1. The blue cline indicates the carbon profile for protein and the other one as carbon profile of ligand bound protein. For instance the blue colour at the bottom line–Zero icod are the region of interest for the ligand design and successful delivery of drug that alter the disease mechanism where all other probes fail to monitor and control. As a matter of point here the probe binds in one place but alters ICOD at stretch of 146–153 decisively. ICOD does play a role in protein alteration naturally along with amino acids that are adjacent to active region and of probe binding place. Alteration of amino acids such as aromatic amino acids at this site alter all the other COD formation that actually alter the COD formation by probe. One can go on testing these kinds of amino acid domain that can alter the due course of action in biology. Alternatively another probe that may be suitable for producing ICOD at these points may be interest to synthetic chemist. All of them are according to the principle of carbon value, true to the carbon domain in the vicinity of molecular interaction where all others fail to explain probe binding. No matter whatever may be carbon plays an important role in probe fixing and accelerate the other biological function of living life.
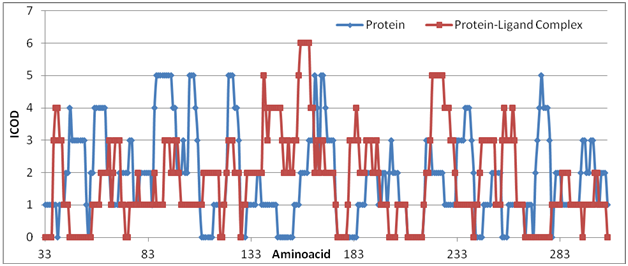
Figure 1 The probe stabilizes protein (at 146–153) with change in carbon domain. All along the amino acids are carbon domain and the zero cod are considered to be active regions.
Another site at 208–216 is of interest for futuristic application of probe binding accordingly. Alternatively active region can also be suppressed at this site of interest. Another at 174–184 is fairly covered with existing probe. All other sites are already occupied by carbon domain value verifiable from blue line. Effective dark brown line of probe bound carbon domain graph for complex indicates the entire carbon deficient sites including 45–55 and 266–278 for further on binding force. Otherwise other site 208–216 is available in both bound and unbound state of protein. Engineering these sites is biologically important for futuristic application of carbon role of binding. Analysis of ICOD for the protein ligand interaction is one of the topics to be considered for treating diseases. There are analyses treating elemental level for drug discovery which again fails in accurate production. However ICOD analysis promisingly increases its strength day by day improving the future of molecular level interaction in the drug discovery platform. Every now and then there is adequate complex formation in the drug discovery profile. Very few leads to success in animal side of research for potential probe finding. Every drug discovery leading to potential probe for diseases are to be considered from bottom of ICOD analysis which are considered here as an address point for initiating probe finding. Every now and then there is going to be addressing probe analysis for securing active regions and the ligand bound state. Active regions are needlessly the binding domain of probe fixing which are demonstrated here again. Active regions are responding to the probe binding but adequately distant enough where all other forces are not influencing
The role of carbon in protein–probe interaction is analysed here. Details on how and where the protein changes in response to probe–protein complex formation are revealed here. Identification and development of active sites can be better exploited from carbon domain formation. Analysis of active site can be utilized for newly devisable probe of action.
None.
None.
The author declares there is no conflicts of interest.

©2019 Ekambaram, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.