International Journal of
eISSN: 2573-2889


Research Article Volume 4 Issue 3
1Department of Chemistry, VSB Engineering College, India
2Kazan Federal University, Russia
Correspondence: Rajasekaran Ekambaram, Department of Chemistry,V.S.B. Engineering College, Karur–639111, Tamil Nadu, India, Tel +91 9524825876
Received: April 26, 2019 | Published: May 6, 2019
Citation: Ekambaram R, Rajasekaran I. Who power sickle cell disease: carbon domain analysis tells all because of design in protein 3d arbitrary internal carbon domain (cod) arrangement. Int J Mol Biol Open Access. 2019;4(3):85–88. DOI: 10.15406/ijmboa.2019.04.00102
Carbon distribution in macro molecules becomes important to address the diseases in living systems. CARd3D program locate these carbon contents in 3D structures of proteins. Here the sickle cell anemia causing mutation in hemoglobin is analysed for betterment of disease cause and compared. Results reveal the involvement of carbon content in disease mutation which is causing sickle cell anemia and identified possible location of protein for salvation.
Keywords: Sickle, COD, carbon-high, carbon domain, anemia, hemoglobin, CARd, role of carbon
Several destructive diseases posing danger to the human life form (example, Ebola virus disease-EVD). Sickle cell anemia, one such disease arises from hereditary disorder. This can cause pain called sickle crises and symptoms like delayed growth and jaundice. Sickle cell disease prevents oxygen from reaching various organs of the body and causing a lot of cell death. Due to this reason, the patients get frequent infections and ultimately leading to death. The reduction in hemoglobin due to short life of the sickle cell causes anemia. The hemoglobin molecule has three components, heme group, an alpha domain and a beta domain. Patients with sickle cell disease have a mutation in the beta domain. As a result of mutation, the RBC takes a concave shape. Eaton and coworkers conclude that sickle RBC adhere to vascular endothelial cells in vitro, perhaps caused by a calcium-induced aberration of membrane topography. This adherence may be a pathogenetic factor in the microvascular occlusions characteristic of sickle cell disease.1 Verification of these adhesion possibly be the carbon role and its structural phenomena that ought to be the central point of focusing. Recently I have reported that globular proteins prefer to have 31.45% of carbon for its stability and can be used as standard for carbon distribution analysis.2–4 Allotment of carbon is responsible for disorders in proteins.5 Hydrophobic interaction is the dominant force crucial for folding.4 The content and distribution of this carbon make a protein to fold in some fashion to have a specific function. Disease proteins are expectedly differing from this carbon distribution profile.6,7 The disorders are mainly due to carbon role in different phenomena may be of evolutionary reason. Toxicity also depends on this factor of carbon role.8 CARd3D program4 exploits the availability of protein 3D structures and reveals the facts of carbon role in macromolecular folding and binding. It can play a crucial role in understanding diseases at molecular level and giving solutions in solving them. Though it is designed for globular proteins, it can be extended further for macromolecular interactions and others as well.9,10
The carbon role in disease mutation is studied here by taking hemoglobin as case study which is crucial for disease salvation and all about carbon domain with emphasis on probe finding in control of the disease where mechanism is understood properly. Newly found carbon domain is expectedly play a crucial role for all human related diseases and salvation. Fact of domain needs11 to be appreciated at all level of molecular disease control mechanism from nano to macro. Macro may be a appreciable one but nano needs to be addressed for global understanding of macro molecular force existence.
Data
The sequence and structure of hemoglobin both in normal and diseased state are retrieved from PDB.12 Protein IDs are 1J3Z (HbA hemoglobin) and 2HBS (HbS hemoglobin). Both contain subunits of alpha and beta globin. Only the beta globin is taken for CARd analysis further because of the mutation that leads to disease is observed in here. Hydrogen atom are added for the X-ray structure using molprobity13 program made available freely online.
Sequence analysis
CARd analysis on the protein sequences are carried on using well known program called CARd. A carbon distribution profile is obtained for different outer lengths. Details of calculation can be observed.2 A plot of carbon profile at amino acid positions is observed. So the carbon profile value obtained gives the idea of location of Globular Amphipathic Domain (GAD), carbon-high hydrophobic regions and reduced-carbon hydrophilic regions.
Structure analysis
CARd analysis using protein sequence gives the GAD and hydrophobic regions. The same thing can be observed using 3D structure of the protein. CARd3D program can be utilized for this purpose. Details again given elsewhere else. Modified CARd3D program capture the Internal Carbon Optimised Domain (COD), which is being equivalent to GAD. The calculation is carried out only at 16Å which capture the phenomena of COD value. Bond length of all possible bonds are analysed for comparison. The protein 3D structure is analysed in particular with bond length of all back bone bonds (namely N-CA, CA-C, C-N, N-H and C=O). For better comparison the bond values of 5 amino acids are group averaged. Change in bond length is plotted for each amino acid position (see figure for CA-C bond). The difference in bond length in normal and sickle cell is plotted for comparison.
Structure analysis
The stable domain becomes reactive in sickle cell by single mutation that take place elewhere in the protein architecture. Cohessiveness explains the disease. Figure 1 shows the variation of COD value along the amino acid sequence. Note that the mutation (E6V) has occurred at position 6 and changes felt beyond 100 amino acids. That the carbon role in COD formation is realised here after some point. Cohessive force coming from far behind is understood well in advance with carbon role using CARd analysis program. Also note that the changes are observed at dia16 and not any other dias. The cohessiveness responds at 16Å. Though one can go on with other dias overall changes shown Figure 2, the physical changes observed based at dia16 including bondlength (Figure 3). The drastic difference in COD value at 92-117 is altered by heme group (result not shown here) both in normal and sickle cell. However the difference is opposite that is the heme reduces the COD value in normal while it increases in sickle cell. This is because of cohesveness of carbon role. Even after this changes there are nonCOD regions at this site that leads to polymerisation that causes sickle cell disease. Comparing the overall changes in COD values for lower dias including D8 upto D15 reveal that not much change in the affinity check whereas at 31-37 and 42-46 the COD is zero meaning that it may be binding elsewhere that happened in polymerisation of haemoglobin chains based on beta globin.
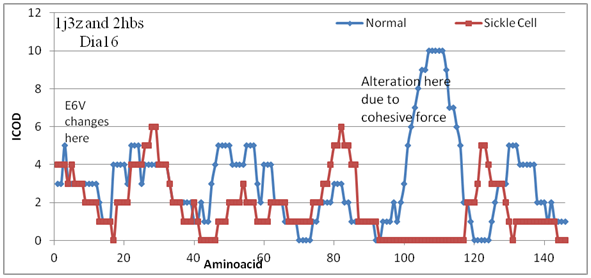
Figure 1 COD changes in beta globin protein without heme in sickle cell. Observe the drastic change at 92-117.

Figure 3 COD changes can be checked by backbone bond length alteration. Note that there is increase in bond length of probable active site at 99-103 considerably.
The bond length variation can be better visualised with COD change in both normal and sickle cell.The result of COD changes in presence of heme is not shown but discussed here. For example 99-103 the COD region in normal beta globin protein becomes nonCOD region where the bond length increased notably. On the other hand there is reduction or nonCOD formation at 44-51 which yielded a negative effectoin bond length. That is to say that the bond counter productive in variation where the polymeriastion is not favorably supported. Notably the nonCOD portion 120-124 in normal protein become COD which is captured by bond length value, that is decreased. Note that the bond length variation is observed for CA-C bond only here as it reflects neatly with change in COD variation. Otherwise other back bone bonds are also captured and compared. There are other effects like double bond character, terminal atoms and hetero atoms come into play. The results are not shown. CA-C alone is good enough to reflect the COD value since it is a C-C bond that occomodate cohessiveness of carbon value. Side chain cannot be compared as it differ from one another, not a common feature here for COD is considered. Uncommon bonds are not considered here. Common for all amino acids are weighted here for COD measure which is C-C of back bone atoms. Otherwise the C-N bond is partial double bond which increase in length on COD formation due delocatiosation of π electrons, counter to C-C bond which may used in opposite manner of COD evaluation. Average of all back bone bond lengths may be not effective in COD valuation. Individual back bone bonds need to be checked carefully for COD valuation.
Note also that the COD reduces the bond lengths due adduct like structure formation. Everywhere except nonCOD regions the bond lengths altered. Normal COD structured proteins have smaller compact structure compared to non adherent protein with random potential of COD formation. Failure to form COD may lead adherent externally which attract external molecule to satisfy them which we call it as active region. Failure here the COD region and not carbon itself forming domain other atoms aswell involved in where one can look into for probe design. Probe design is one of the challenging activity in bioinformatics forum to be involved in where COD comes into play. Overall growth in industry that might be estimatted to be huge in size where as COD will solve it in no time naturally. Here it is estimatted for sickle cell disease is huge in response overall growth of humanity based in Africa. Naturally one might be thinking this way to solve the long standing probe finding to disease control elsewhere in Indian market. Probability of amino acid stretches in beta globin of normal and sickle cell reveal that the changes are not at smaller circle but slightly larger. It identifies stretch 34-43 as less probable one. In other calculations it is found out to be hydrophobic stretch which is less probable. The stretch 116-119 is also less probable and hydrophobic site which is found out to be in the interface of dimer alpha-beta. This is responsible for alpha-beta dimerisation, confirmed by COD calculation.
Sequence analysis
Based on sequence information, CARd program using outer length of 108 analyses and finds 32-48, 70-75, 96-103 and 114-127 are hydrophobic stretches. These are the potential sites of active regions where the dimerisation, drug binding or polymerization can take place. From probable analysis 116-119 has been identified as less probable site. Further crystal structure visualization identifies this site involved in dimerisation of A and B chains and probably not available for drug binding. Apart from these sites the other regions are identified as GAD regions and self satisfied one. Alpha globin analysed by CARd is able locate the hydrophobic stretches as follows 20-27, 31-36, 37-47, 93-98, and 111-120. Further look into probability analysis and flat side chain amino acid identification identifies 40-47 as potential site of interest. Histidine is one of the amino acid with planar aromatic side chain which is responsible for activity in most of the proteins, particlarly here in globin. How will the mutation in beta globin affect COD in alpha globin? The change in COD at dia16 is computed for comparison in both normal and sickle cell. Due to the cohessiveness, it varies in COD stretch length but position remain same. Remarkably at stretch length 19-25, there is change from nonCOD to COD during sickle cell disease. Worthy that the cohessiveness of adjoining domain of proteins alter the active site of A chain.
A structure of COD comprise of amino acids combination that form appropriae carbon distribution. They are made from carbon and polar atoms of those amino acids organised in some facts of form. There are varieties of COD forming compositions evolved which have been adapted to a broad range of stability in proteins. Where ever it occurs in proteins, they are subjected to water repellent. That is the linings of amino acids in COD build to water shield. The COD stretch that has a high COD command is enough to maintain stability. All amino acids used in the forming of COD are important for water shield in biological conditions. Based on the application, CODs must resist molecule of interaction, oppose chemical reaction, catalytic action and similar one. As variety of composition of CODs instruct a variety of performance and properties, many CODs have been evolved for different stability purposes. But here in beta globin the COD becomes non COD upon mutation which aggregate the haemoglobin to damage regular RBC to sickle cell that ultimately damage the whole functioning of other system in the body.
The COD forming bond length of selected back bone atoms are observed to be reduced in nature of all time. The internal amino acids involved in carbon domain form a compact structure. The atoms are facing each other in such ways that meet the optimum value of carbon all over structure. The arrangement is to share electrons among them. The bond lengths reduced due to this in the structure. The adduct structure formed by this is behaving like as if aromatic compound that can be water repellent. This can oppose alteration by any other interacting atoms in biological condition. Better visualization of COD is performed based on bond length of back bone atoms such as N-CA, CA-C, C-N, N-H and C=O. Due to partial double bond, terminal atoms and hetero atoms, except CA-C bond all other bonds are not considered in COD evaluation. But CA-C bond value expresses neatly the phenomena of COD as well as performing and non performing regions in proteins. Overall good results agreeable to COD values are noticed. Performing individual bond of all back bone atoms can reveal exclusivity of COD pattern. Performance based on bond length can be better exploited in due course of action. Overall nice development based in bond length is hoped to help in the near future either in drug discovery or stabilization phenomena that alter protein to go functional in increased activity. One may try with redevelopment of protein with higher stability and activity using this.
CARd3D program able to explain the defects due to mutation. Carbon domains are answerable for sickle cell anemia. Identify a drug that recognizes the polymerization site (probably 99-103) in beta globin to stop sickle formation.
None.
Author declares that there is no conflicts of interest.

©2019 Ekambaram, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.