International Journal of
eISSN: 2381-1803


Research Article Volume 8 Issue 2
1Departments of Nursing, Hengdian Wen Rong Hospital, China
2Department of Physical examination center, Hengdian Wen Rong Hospital, China
3Biotecan Medical Diagnostics Co., China
Correspondence: Rilei Gao, Department of Physical examination center, Hengdian Wen Rong Hospital, Hengdian, China, Tel 86-0579-89301686
Received: July 17, 2017 | Published: July 31, 2017
Citation: Jin Y, Shen L, Gao R, Dong S (2017) Early Diagnosis with 11-Gene Panel Screening for Non Small Cell Lung Cancer. Int J Complement Alt Med 8(2): 00254. DOI: 10.15406/ijcam.2017.08.00254
NSCLC, caused by various mutations in a spectrum of cancer driver genes, could be early diagnosed by genetic screening and may have corresponding drug responses. In an effort to complete the clinical application of precision diagnosis in lung cancer, increase the confirmed diagnostic rate and also provide the reference and inspiration for precision diagnosis of other tumors. We designed a panel of 11 genes, BRAF, EGFR, KRAS, HRAS, IDH, PIK3CA, RET, PTEN, C-KIT, SMAD4 and HER2, which played important roles in the progression of lung cancer to help diagnosis lung cancer. The results showed that compared with the healthy controls, the concentrations of BRAF, EGFR, KRAS, HRAS, IDH, PIK3CA, PTEN, C-KIT, SMAD4 and HER2 were higher in patients with benign lesions and lung cancer. There were significant difference between healthy controls and patients with lung cancer, also between patients with benign lesion and patients with lung cancer in the concentration of BRAF, EGFR, KRAS, HRAS, IDH, PIK3CA, C-KIT, SMAD4 and HER2, and there was no difference between healthy controls and patients with benign lesion. ROC analysis was further performed to evaluate the diagnosis efficiency. Sensitivity and Specificity of BRAF, EGFR, KRAS, IDH and SMAD4 was much higher and with the combination of these genes, that is the panel, the sensitivity was 100%. The area under the curve of BRAF, EGFR, KRAS, IDH, SMAD4 and the panel was 0.786, 0.803, 0.87, 0.906, 0.887 and 1.000 respectively. The panel of these 11 genes is of great clinical value for diagnosis of lung cancer and target therapy for that it expanded the coverage of mutated gene and improved the diagnostic efficiency.
Keywords: NSCLC, non small cell lung cancer, early diagnosis, gene panel
Lung cancer is a heterogeneous disease with the symptoms and signs of cough, hemoptysis, chest pain, and fever and dyspnea chest discomfort and is classified into small cell (SCLC) and non-small cell (NSCLC) lung cancers.1 NSCLC is the most common histological types of lung cancer, accounting for about 80-85% of all the types and can be further classified into squamous cell carcinoma, adenocarcinoma and large Cell carcinoma.
Despite advances in medicine, lung cancer remains the leading cause of cancer-related mortality. More than 50% of lung cancer patients are diagnosed with metastatic disease and have a 5-y survival rate of <5%.2 Based on the past data, the estimated number of newly diagnosed lung cancer cases is expected to reach 221,200, with the number of deaths cause by lung cancer predicted to reach 158,040, accounting for 27% of all cancer-related deaths.3
National Institute for Health and Clinical Excellence (NICE) guidelines recommend radical surgery for stage I or II NSCLC. Chemotherapy and radiotherapy are recommended for later-stage NSCLC patients.4 Other treatment interventions including chemotherapy, radiotherapy, immunotherapy and targeted therapy have also been shown to improve lung cancer survival.5 Among those treatments, targeted therapy aimed at mutated genes. And the test of gene expression in precision medicine worked as guidelines of targeted therapy. Therapeutic options for patients diagnosed with lung cancer have significant growed due in major part to improved technological ability of diagnosis.6,7 Early stage diagnosis and surgery of NSCLC patients could result in a 5-year survival rate of up to 55–80%.8 Therefore, one of the major challenges for lung cancer lies in early precision diagnosis. Currently, routine clinical diagnostic methods includes tumor markers detection, computed tomography screening, histopathological detection and so on which are basically from the point of serology, imaging and pathology.9 and the optimal means of diagnosis are uncertain and lack of data of precision medicine owing to the involvement of different tumors, patient and health-care factors, often in combination.10 Numerous genetic alterations associated with lung cancer have been identified that might contribute to the development of specific types of lung cancer, patterns of metastasis, drug resistance, or disease recurrence.11 To test and fully understand the related genes involved in the development of lung cancer will further help the diagnosis much more clear and therapy much more targeted. The precision diagnosis related researches in lung cancer and other cancers mainly referred to drive genes, heterogeneity, sub-classification,12,13 and researches about liquid biopsy. A series of genes were reported to abnormal expressed in non-small cell lung carcinomas (NSCLCs) including epidermal growth factor receptor (EGFR) which more than 60% of NSCLCs expressed.14 The test of these mutated genes could be performed with blood samples with the development of liquid biopsy which make the test of gene expression convenient and repeatable.15 There's also the development of new technology, such as fluorescent chemical probes, which aid the tumor diagnosis and therapy.16
Most studies still remain preclinical or in the stage of research, so in the present study, a panel of 11 genes was designed to verify the results of previous studies in this area. The test of the panel of these genes will complete the clinical application of precision diagnosis in lung cancer, increase the confirmed diagnostic rate which is of great significance for clinical diagnosis and therapy and also provided the reference and inspiration for precision diagnosis of other tumors.
Study participating
At Hengdian Wen Rong hospital, we recruited patients diagnosed with NSCLC, benign lesions including tuberculosis, pneumonia and inflammatory pseudo tumor and so on and healthy persons based on the physical and laboratory examination, male or female, 55 to 74 years of age. The study was approved by the institutional review board of the hospital and all participants provided written informed consent before participation.
Screening equipment and procedures
According to the related researches and clinical situation, we designed a panel contained 11 genes which played important roles in the progression of lung cancer to help diagnosis lung cancer. The 11 genes were as follows: BRAF, EGFR, KRAS, HRAS, IDH, PIK3CA, RET, PTEN, C-KIT, SMAD4 and HER2. Plasma samples were collected at early morning from all participants, and other clinical data was also recorded which was shown in Table 1 & 2.
|
Characteristic |
Healthy Control N (%) |
Benign Lesion |
Lung Cancer |
|
Gender |
|
|
|
|
Male |
22 (52.4%) |
19 (82.6%) |
10 (52.6%) |
|
Female |
20 (47.6%) |
4 (17.4%) |
9 (47.4%) |
|
Age |
|
|
|
|
40-55yr |
12 (28.6%) |
15 (65.2%) |
5 (26.3%) |
|
56-70yr |
27 (64.3%) |
7 (30.4%) |
11 (57.9%) |
|
≥71yr |
3 (7.1%) |
1 (4.3%) |
3 (15.8%) |
Table 1 Demographics of the study population
DNA extraction
Plasma was separated via centrifugation at 800 g for 10 minutes, transferred to micro centrifuge tubes and centrifuged at 16,000g for 10 minutes to remove cell debris. Circulating DNA was isolated from 1–5 ml of plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen). Briefly, buffer ACL was added into plasma and incubated in 60℃, 30 min, and then buffer ACB was added and incubated in 4℃, 5 min. Then mixture was separated by HiPure CFDNA Mini column, and eluted by buffer TE. DNA concentration and purity were quantified using a Qubit 2.0 Fluor meter (Life Technologies) and an Agilent 2100 Bioanalyzer (Agilent Technologies).
CtDNA mutation detection via ddPCR
The mutation levels of ctDNA were measured using a ddPCR system (Bio-Rad). Briefly, primers and probes for detection of BRAF, EGFR, KRAS, HRAS, IDH, PIK3CA, RET, PTEN, C-KIT, SMAD4 and HER2 were obtained following the recommended protocol (Bio-Rad Laboratories). The PCR mixture was loaded into an 8-channel, single-use consumable droplet generation cartridge. Approximately 20,000 mono-dispersed droplets for each sample were prepared using the Quanta Life droplet generator. The water-in-oil emulsions were pipette-transferred to a 96-well PCR plate and subjected to amplification cycles. (Cycling protocol: 95 °C for 10 min, 40 cycles of 94 °C for 30 s and 60 °C for 60 s, and a final step at 98 °C for 10 min). Then plates were loaded into the commercially obtained QX100 droplet reader (Bio-Rad). And the concentrations (copy numbers) of the targets in the samples were determined using QuantaSoft software. The threshold of diagnosis was determined according to the results of the healthy controls. When concentrations (copy numbers) of the targets were above the threshold, the sample was considered to be positive.
Statistical analysis
Gender and age of the participants, type of disease, histological features and disease stage of patients were statistically described. Positive rate of diagnosis was the proportion of the patients with gene mutations. Thresholds for gene mutations were as follows: BRAF, 0-1; EGFR, 0-1.36; KRAS, 0-2.07; HRAS, 0-0.24; IDH, 0-0.79; PIK3CA, 0-0.31; RET, 0-0.26; PTEN, 0-0.38; C-KIT, 0-0.9; SMAD4, 0-0.77; HER2, 0-0.78. The continuous data of ddPCR in the three groups was compared using one-way ANOVA. Logistic regression was performed to integrate the 11 genes and calculate the data of the panel. ROC curve was performed to test the diagnostic efficiency of the panel and each gene. P<0.05 and p<0.01 were considered to be statistically significant and highly significant difference, respectively. All statistical analyses were performed using the SPSS version 16.0 software package.
Recruitment and screening
A total of 23 benign lesions cases, 19 lung cancer patients and 42 healthy controls were included in the study. All of them were recruited between August 2016 and December 2016 from Hengdian Wen Rong hospital. Clinical characteristics of the study population have been shown in table 1 and 2. Benign lesions including tuberculosis 3 cases, pneumonia 10 cases, inflammatory pseudo tumor 7 cases and lung texture enlargement 3 cases. Of the patients with lung cancer, 13 patients were adenocarcinoma, 6 were squamous-cell carcinoma and according to the TNM stage, number of patients of stage I, II, III and IV was 11, 1, 5 and 2 respectively.
|
Histologic features and stage |
Lung cancer |
|
Histologic features |
|
|
Adenocarcinoma |
13 |
|
Squamous-cell carcinoma |
6 |
|
TNM Stage |
|
|
I |
11 |
|
II |
1 |
|
III |
5 |
|
IV |
2 |
Table 2 Characteristics of the patients with lung cancer
We performed lung cancer detection with the panel of genes in all the participants and calculated the positive rate which was shown in Table 3. In healthy controls and patients with benign lesion, no positive mutated genes were detected while in patients with lung cancer, at least one mutated gene was screened (the positive rate of the panel=100%) and the positive rate was above 50% in EGFR, IDH, SMAD4.
|
Group |
Positive (%) |
|||||||||||
|
BRAF |
EGFR |
KRAS |
HRAS |
IDH |
PIK3CA |
RET |
PTEN |
C-KIT |
SMAD4 |
HER2 |
Panel |
|
|
Healthy control |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Benign |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Lung |
5/19 |
10/19 |
9/19 |
2/19 |
10/19 |
2/19 |
0 |
0 |
8/19 |
10/19 |
5/19 |
19/19 |
Table 3 Diagnostic rate of the panel and each gene in each group
Gene concentration of the panel
We compared the concentrations (copy numbers) of the 11 genes among the three groups to highlight the differences. Compared with the healthy controls, the concentrations of BRAF, EGFR, KRAS, HRAS, IDH, PIK3CA, PTEN, C-KIT, SMAD4 and HER2 were higher in patients with benign lesions and lung cancer. There were significant difference between healthy controls and patients with lung cancer, also between patients with benign lesion and patients with lung cancer in the concentration of BRAF, EGFR, KRAS, HRAS, IDH, PIK3CA, C-KIT, SMAD4 and HER2, and there was no difference between healthy controls and patients with benign lesion. In the test, the concentration of RET is lower in patients compared with the healthy controls because of less patients with RET mutation resulting in the mean concentration was low in patients (Figure 1).
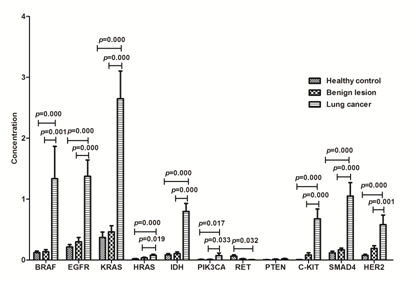
Figure 1 Concentration (copy numbers) of the 11 genes in the panel among healthy controls, patients with benign lesion and lung cancer. Healthy controls (n=42); patients with benign lesion (n=23); patients with lung cancer (n=19).
Diagnostic efficiency of the panel
We performed logistic regression to integrated the concentration of the 11 genes and the result showed that in this panel we designed, EGFR, HRAS, IDH, PIK3CA and C-KIT were of much more significance in the panel (the results of last step of logistic regression was shown in Table 4) and other genes may play a reference role.
|
Variable(s) |
B |
S.E. |
Wals |
df |
Sig. |
Exp(B) |
|
EGFR |
50.426 |
4611.309 |
0.000 |
1 |
0.991 |
7.941E21 |
|
HRAS |
259.239 |
29852.964 |
0.000 |
1 |
0.993 |
3.855E112 |
|
IDH |
135.161 |
10084.857 |
0.000 |
1 |
0.989 |
5.006E58 |
|
PIK3CA |
116.703 |
13720.328 |
0.000 |
1 |
0.993 |
4.824E50 |
|
C-KIT |
150.665 |
11434.265 |
0.000 |
1 |
0.989 |
2.709E65 |
|
Constant |
-119.732 |
8665.931 |
0.000 |
1 |
0.989 |
0.000 |
Table 4 Key parameters in logistic regression
ROC analysis was further performed to evaluate the diagnosis efficiency. Sensitivity and specificity of BRAF, EGFR, KRAS, IDH and SMAD4 was much higher and with the combination of these genes, that is the panel, the sensitivity was 100% (Figure 2). Area under the curve was also calculated (Table 5) and the area above 0.78 was considered to be significant and could be used for clinical diagnosis. The area under the curve of BRAF, EGFR, KRAS, IDH, SMAD4 and the panel was 0.786, 0.803, 0.87, 0.906, 0.887 and 1.000 respectively.
|
Test result variable(s) |
Area |
Std. Errora |
Asymptotic Sig.b |
Asymptotic 95% confidence interval |
|
|
Lower Bound |
Upper Bound |
||||
|
BRAF |
0.786 |
0.072 |
0.002 |
0.645 |
0.927 |
|
EGFR |
0.803 |
0.072 |
0.001 |
0.662 |
0.945 |
|
KRAS |
0.87 |
0.062 |
0.000 |
0.749 |
0.990 |
|
HRAS |
0.658 |
0.088 |
0.081 |
0.485 |
0.830 |
|
IDH |
0.906 |
0.05 |
0.000 |
0.807 |
1.000 |
|
PIK3CA |
0.566 |
0.091 |
0.464 |
0.389 |
0.744 |
|
RET |
0.418 |
0.088 |
0.363 |
0.245 |
0.591 |
|
PTEN |
0.562 |
0.091 |
0.495 |
0.384 |
0.74 |
|
C-KIT |
0.737 |
0.081 |
0.009 |
0.578 |
0.896 |
|
SMAD4 |
0.887 |
0.057 |
0.000 |
0.775 |
0.999 |
|
HER2 |
0.706 |
0.082 |
0.023 |
0.546 |
0.866 |
|
Panel |
1.000 |
0.000 |
0.000 |
1.000 |
1.000 |
Table 5 Area under the Curve
A: Under the nonparametric assumption
B: Null hypothesis: true area=0.5
NSCLC, caused by various mutations in a spectrum of cancer driver genes, may have corresponding drug responses. Genetic screening is required for early diagnosis and the design of customized therapies to improve patient outcomes.17
Oncogenic mutations in the epidermal growth factor receptor (EGFR) are found in a subset of patients with non–small cell lung cancer (NSCLC) and serve as important predictive biomarkers in this disease.18-20 Another high prevalence of mutation is PIK3CA, which mutated in both adenocarcinomas and squamous cell carcinomas.21 RET is associated with the development of NSCLC, Several RET rearrangements, specifically fusions, have been identified in NSCLC.22,23 IDH1, an isotype of IDH, was reported to be significantly up regulated in lung cancer and was proposed to be a putative biomarker for the diagnosis of NSCLC.24 Besides, HRAS.25 and PTEN.26 also were reported that they could be used for identifying patients with NSCLC for their frequently mutations and activation in the process of NSCLC.
Molecular detection in lung cancer also was used as predictor, such as TGF-β1, which may be an independent predictor of survival in resected lung adenocarcinoma patients.27 SMAD4 was identified as a tumor suppressor gene and was associated with tumor invasion, metastasis and prognosis in different cancers including NSCLC.28,29 C-Kit have critical roles in cell proliferation and differentiation in patients with NSCLC which indicate that C-Kit is a strong, independent prognostic factor in NSCLC.30
KRAS frequency appears to be similar in primary and central nervous system metastases and the mutations are associated with tumor resistance to EGFR TKIs (erlotinib, gefitinib) and to monoclonal antibody against EGFR (cetuximab). Although BRAF mutation is rare in patients with NSCLC, and its presence is associated with sensitivity of tumor cells to BRAF inhibitors (vemurafenib, dabrafenib). So the detedtion of KRAS and BRAF could be used not only as biomarkers of NSCLC, but also as guides of target therapy.31 HER2 is one of the driver mutations in lung cancer and is found in about 2% of lung adenocarcinomas. Although some gene mutations were rare in lung cancer, the detection of these mutations actually leads to precise molecular-targeted therapies.32
As reported, assessing the mutation of EGFR.33 and the other genes were the accepted standard of care worldwide for patients with lung cancer.
To increase the diagnosis efficiency and screen the related oncogenic gene at one time conveniently, we developed a gene panel integrating all these highly-mutated genes together based on all the researches above.
In our study, the concentration of genes in the panel were significantly higher in patients with lung cancer except RET, PTEN because of the few participants. ROC analysis showed that the panel of 11 genes was of high clinical sensitivity and specificity. All the results proved that the panel could be used to distinguish patients with lung cancer from healthy controls.
Coincidentally, a NextDaySeq lung panel of EGFR, KRAS, PIK3CA and BRAF designed by other team exhibited good clinical performance, strongly supporting the implementation of gene mutation assay in routine clinical use to facilitate therapeutic decision-making for non-small cell lung cancer patients.34. Our panel added seven more important genes related to NSCLC to improve the diagnosis effect further. With this improvement, the panel of 11 genes could be used to help diagnosis, noninvasive, convenient and without side effects compared with CT (Computed Tomography), MRI (Magnetic Resonance Imaging) and chest radiograph. However, there was some small flaws with this panel, that was, not all the genes would mutated in all patients for some of the genes were not frequency mutated in NSCLC. As a result, the combination of these 11 genes in one panel is necessary for that it expanded the coverage of mutated gene and improved the diagnostic efficiency.
The panel of 11 genes has the potential to be used for diagnosis of lung cancer with high sensitivity and specificity. Among the 11 genes, BRAF, EGFR, KRAS, IDH and SMAD4 played a greater role in the panel and the combination of 11 genes could improve the diagnosis efficiency.
None.
There is no conflict of interest.
None.

©2017 Jin, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.