International Journal of
eISSN: 2574-9862


Research Note Volume 2 Issue 3
1Department of Preventive Veterinary Medicine and Animal Health, Sao Paulo University, Brazil
2Department of Veterinary Medicine, Sao Paulo University, Brazil
Correspondence: Pedro Henrique Magalhaes Cardoso, Rua Professor Orlando Marques Paiva, Sao Paulo University, Postal Code: 05508270, Brazil
Received: June 24, 2017 | Published: October 3, 2017
Citation: Cardoso PHM, Maganha SRL, Sousa RLM, et al. First report of megalocytivirus in red piranhas (pygocentrus nattereri) by molecular diagnosis in Brazil. Int J Avian & Wildlife Biol. 2017;2(3):82-84. DOI: 10.15406/ijawb.2017.02.00023
The genus Megalocytivirus belongs to the family Iridoviridae and due to the high mortality rates among infected animals, represents a significant threat to world aquaculture. The affected fish present general and nonspecific symptoms, which makes the early diagnosis of the infection difficult. Nine samples from ornamental fish of the species Poecilia reticulata and Pygocentrus nattereri with nonspecific symptomatology were submitted to molecular diagnosis for Megalocytivirus through the PCR technique. Infection by this genus was detected in 100% of the analyzed samples, and three of them were sent for sequencing. All samples showed identity values of 100% with the Megalocytivirus Sabah/RAA1/2012 strain BMGIV48 described in Malaysia. These results indicate, for the first time, the circulation of Megalocytivirus among Brazilian ornamental fish, which emphasizes the necessity of larger studies regarding this important viral genus.
Keywords: megalocytivirus, ornamental fish, live fish foood, molecular diagnosis
MCP, major capsid protein; ISKNV, infectious spleen and kidney necrosis virus; FAPESP, fundacao de amparo a pesquisa do estado de sao paulo
The family Iridoviridae is composed of five genera that affect a wide variety of animal species, including freshwater and saltwater fish and invertebrates. Two genera, Iridovirus and Chloriridovirus, contain viruses of invertebrates, while three genera, Ranavirus, Megalocytivirusand Lymphocystivirus, contain viruses that infect lower vertebrates. The genera Megalocytivirus and Ranavirus are emerging pathogens of global distribution that cause systemic diseases of high severity. Ranaviruses are also important amphibian pathogens. The genus Lymphocystivirus causes lesions on the surface of fish epidermis and is rarely responsible for economic losses.1
Infections by Megalocytivirus have been reported in several species of ornamental fish, including Trichogaster leeri, T. microlepis, Colisa lalia, Poecilia sphenopsx, Poecilia reticulata, Astronotus ocellatus, Hyphessobrycon innesi, Pterophyllum scalare,2 Pterapogon kauderni,3 Platax orbiculare,4 Xiphophorus helleri, Aphyosemion gardneri, Xiphophorus maculatus, Poecilia latipinna,5 among others and can cause significant economic losses worldwide.
Clinical signs are nonspecific and resemble the clinical signs observed in many other diseases in fish. Darkened skin, irregular swimming (including rotating movements) or positioning on the surface of the water, increased respiratory movements, distention of the abdominal cavity, ulceration, hemorrhages (including punctate hemorrhages in the skin and gills), pallor of the gills, anemia, erosion of the fins, whitish stools and high mortality rates are the most common clinical signs. Necropsy shows necrosis in many internal organs such as spleen, kidneys, and liver. Other organs and tissues, including muscles, gonads, heart, gills and gastrointestinal tract may also be affected. Some fish may have visible hemorrhagic fluid in the body cavity.4,6 The aim of the study was identify red piranhas (Pygocentrus nattereri) used as ornamental fish suspected of viral infection by molecular diagnosis, and to alert to the presence of Megalocytivirus in Brazil.
Fifty red piranhas (Pygocentrus nattereri), originating from the Amazon basin through extractive fishery and acquired by a wholesaler of ornamental fish in Sao Paulo, showed nonspecific clinical signs after being fed with live fish food (Poecilia reticulata). Clinical signs included loss of appetite, irregular swimming and positioning on the surface of the water (Figure 1a), lethargy and respiratory movements that started three days after feeding with live fish, which progressed to 94% mortality within five days after the onset of clinical signs.
Five specimens of red piranhas (Pygocentrus nattereri) were sacrificed and necropsied (Figure 1b). At necropsy, hemorrhagic points in the organs were observed, more evident in the gastrointestinal tract (Figure 1c) and gills (Figure 1d) observed under 4X microscopy. On suspicion that live fish food was the source of the problem, immediately, the responsible veterinarian sent a total of eight samples of red piranhas (Pygocentrus nattereri) and a batch of approximately 30 frozen guppies fish (Poecilia reticulata) to the Laboratory of Zootechnical Hygiene of FZEA-USP for molecular diagnosis through the technique of nested-PCR and nucleotide sequencing in order to identify possible viral agents as cause of these alterations in the animals studied.
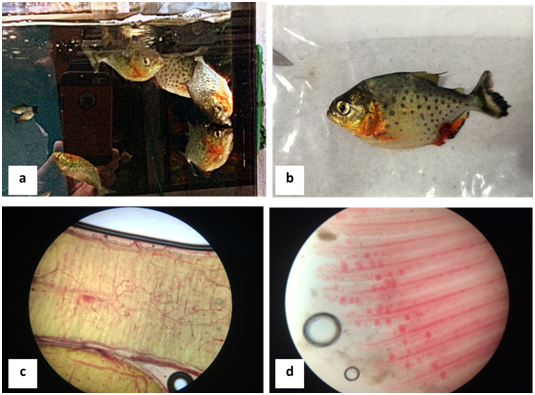
Figure 1 (A) Lethargic piranhas positioned on the surface of the water and with irregular swimming (B) submitted to necropsy (C) hyperemic intestine and (D) gills with hemorrhagic spots.
In the laboratory one specimen of the species Poecilia reticulata and the eight specimens of the species Pygocentrus nattereri were submitted to molecular diagnosis. For the extraction of DNA, fragments of approximately 50 mg of these tissues (pool) were collected; DNA extraction was then performed using the NucleoSpin Extract II kit (Macherey-Nalgel, Germany). The extracted DNA was measured by spectrophotometry (DS-11, DeNovix, USA) according to the absorbance ratio A260/A280. PCR was performed from the DNA extracted for the partial amplification of the Megalocytivirus Viral Major Capsid Protein (MCP) gene. The primers used in nested-PCR are described in Table 1.7
Name |
Orientation |
Sequence (5'-3') |
Genus |
Product (Bp) |
MV-F |
Sense |
ATGTCTGCAATCTCAGGTG |
Megalocytivirus |
1,362a |
MV-R |
Antisense |
TTACAGGATAGGGAAGCCTGC |
Megalocytivirus |
|
nMV-F |
Sense |
CACCGCAACGTGCAAAGCAA |
Megalocytivirus |
369a |
nMV-R |
Antisense |
TTGACTGCAATAACGACCAGTTCAAAC |
Megalocytivirus |
Table 1 Primers used to obtain PCR products fromtheMCP genes of Megalocytivirus.
Samples with band pattern close to that expected in the nested-PCR were excised from the agarose gel, and the DNA fragments contained therein were extracted using the illustra™ GFX™ PCR DNA and Gel Band Purification (GE Healthcare, USA) kit, According to the manufacturer's recommendations. For nucleotide sequencing, the BigDye® Terminator v3.1 Cycle Sequencing Ready Reaction Kit (Applied Biosystems/ Thermo Fisher Scientific, EUA), containing AmpliTaq DNA Polymerase, was used according to the manufacturer's specifications. The BioEdit program, version 7.0.98 was used to edit the sequences while to the search for the similarity of the sequences generated and edited, the BLAST program, version 2.09 was used. Editing and multiple alignments of the nucleotide sequences obtained were performed in the ClustalW program, version 1.4,10 and, implemented in the BioEdit Sequence Alignment Editor program, version 7.0.2.8 Visualization of the alignment, using the deduced amino acid sequences, was performed through the Jalview program, version 2.8.1.11
All the nine processed samples (a sample of Poecilia reticulate and eight samples of Pygocentrus nattereri) were positive for the molecular diagnosis for Megalocytivirus, and 3 of them presented ideal DNA concentration for nucleotide sequencing. BLAST analysis of the 369 bp sequences obtained revealed that 3 of the 2 Brazilian sequences analyzed showed 100% identity with the collinear region of the MCP gene of Megalocytivirus Sabah/RAA1/2012 strain BMGIV48 (GenBank Access Number: JQ253374), originating from Malaysia (Figure 2), and also with the strain described as belonging to the species infectious spleen and kidney necrosis virus (ISKNV), which is one of the species within the genus Megalocytivirus (KX354219). The red piranha is a common native species in the region of the Amazon basin and is in a vulnerable conservation status. This was the first report of molecular diagnosis of Megalocytivirus in Brazil in red piranhas, which draws attention due to the problems that the virus can cause.
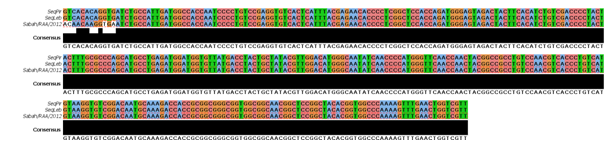
Figure 2 Part of the alignment between the deduced sequences of amino acid sequences of the MCP gene of Megalocytivirus detected in the present study and of the Sabah/RAA1/2012 sequence.
The occurrence of the nonspecific clinical signs3,4,6,12 in red piranhas after feeding with live fish, associated with the findings in the necropsy of organ hemorrhage, have led to the suspicion of diseases of viral etiology. Further studies are needed to confirm whether fish used as food were a source of Megalocytivirus infection for native species. In this study, the Megalocytivirus was detected in a sample of Poecilia reticulata and in the eight samples of Pygocentrus nattereri.
In 2015, Rimmer et al.13 found a positive result in several species of fish. The majority of the animals with positive results in the PCR were apparently healthy and had a low viral load, which indicates that certain species are potential reservoirs of the virus.13
This is the first report of molecular diagnosis in Pygocentrus nattereri in Brazil, suggesting that the native species of the Brazilian basin is susceptible to the virus and alerting us to the presence of the virus in Brazil. To confirm the molecular diagnosis and horizontal transmission by food fish, further and more advanced studies are needed including histopathology and electron microscopy of the samples.
This research was supported by Fundacao de Amparo a Pesquisa do Estado de Sao Paulo (FAPESP, Grant number 2014/04327-7).
The author declares no conflict of interest.

©2017 Cardoso, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.