eISSN: 2469-2778


Literature Review Volume 12 Issue 2
1Cancer and Molecular Medicine Research Group (CAMMO), Columbia
2Los Cobos Medical Center, Columbia
Correspondence: Jheremy S Reyes, Cancer and Molecular Medicine Research Group (CAMMO), Columbia
Received: July 19, 2024 | Published: August 9, 2024
Citation: Reyes JS, Reyes JE, Picón LT, et al. Evaluating the role of pirnas and piwi-like proteins as biomarkers and therapeutic targets in leukemia and lymphoma: a comprehensive systematic review. Hematol Transfus Int. 2024;12(2):45-51. DOI: 10.15406/htij.2024.12.00331
Introduction: This systematic review synthesizes current evidence on the role of PIWI-interacting RNAs (piRNAs) in leukemia and lymphoma, emphasizing their potential as diagnostic markers and therapeutic targets. Leukemia and lymphoma represent significant challenges in oncology, warranting exploration of novel biomarkers beyond conventional methods. piRNAs, a class of small non-coding RNAs, exhibit dysregulated expression in tumor tissues and biological fluids, suggesting their utility in disease diagnosis and targeted therapy.
Methods: A systematic search was conducted in PubMed and ScienceDirect databases following PRISMA guidelines. Eligible studies explored piRNA expression, diagnostic efficacy in leukemia and lymphoma tissues, and therapeutic implications. Included studies spanned from 2000 to 2024 and underwent rigorous quality assessment.
Results: Initially identifying 26 papers, the review included 5 studies meeting inclusion criteria. These studies identified specific piRNAs, such as piR-32877 and piR-30473, with significant diagnostic potential in leukemia and lymphoma (AUC = 0.78). Insights from extracellular vesicle-derived piRNAs, like piR-36225, suggested promising applications for disease monitoring (AUC = 0.69).
Conclusions: piRNAs emerge as promising biomarkers for non-invasive diagnosis and therapeutic targets in leukemia and lymphoma. Future research should validate these findings across diverse populations and elucidate underlying mechanisms to advance clinical utility.
Keywords: PIWI-interacting RNAs, piRNAs, leukemia, lymphoma, biomarkers, therapeutic targets
piRNAs: small RNAs called piwi-interacting RNAs; ALL, acute lymphoblastic leukemia; AML: acute myeloid leukemia; DLBCL, diffuse large b-cell lymphoma; CLL, chronic lymphocytic leukemia
Recent advancements in oncology have increasingly highlighted the intricate roles of small non-coding RNAs, particularly PIWI-interacting RNAs (piRNAs) and PIWI-like proteins, in the pathogenesis and therapeutic strategies of hematologic malignancies such as leukemia and lymphoma. These molecules, once considered primarily as guardians of germline integrity, are now recognized for their multifaceted functions in gene regulation and epigenetic modulation across various cancer types. In the context of leukemia and lymphoma, piRNAs and PIWI-like proteins have emerged as potential biomarkers (genes, gene products, proteins, hormones, or other molecules that are produced by the cancer cells themselves or by other cells in response to the presence of cancer) for disease prognosis and as promising targets for novel therapeutic interventions, and as an alternative to the markers currently used (Table 1), which do not have the expected diagnostic and prognostic performance for these types of pathologies.1–5
|
Disease |
Markers |
Technique |
|
Acute Lymphoblastic Leukemia (ALL) |
CD10, CD19, CD20, CD22, CD79a (B-cell markers); CD3, CD4, CD5, CD7, CD8 (T-cell markers); Tdt (Terminal deoxinucleotidyl transferase) |
Flow cytometry, Immunohistochemistry (IHC) |
|
Acute Myeloid Leukemia (AML) |
CD13, CD33, CD34 (Myeloid markers); MPO (Myeloperoxidase); NPM1, FLT3, CEBPA (genetic mutations) |
Flow cytometry, PCR, Sequencing |
|
Chronic Lymphocytic Leukemia (CLL) |
CD5, CD19, CD20, CD23 (B-cell markers); ZAP-70, CD38 (prognostic markers) |
Flow cytometry, Immunohistochemistry (IHC) |
|
Hodgkin Lymphoma |
CD15, CD30 (Reed-Sternberg cell markers); PAX5 (B-cell marker) |
Immunohistochemistry (IHC) |
|
Non-Hodgkin Lymphomas (NHL) |
CD20 (B-cell marker); CD3, CD4, CD5, CD7, CD8 (T-cell markers); BCL2, BCL6, MYC (genetic alterations) |
Flow cytometry, PCR, Sequencing, FISH |
|
Burkitt Lymphoma |
CD10, CD19, CD20, CD22 (B-cell markers); MYC (t(8;14) translocation involving MYC gene) |
Flow cytometry, FISH, PCR |
Table 1 Key markers used for the diagnosis and prognosis of various leukemias and lymphomas, as well as the techniques employed to detect these markers
This systematic review aims to explore and critically analyze the scientific evidence on the involvement of piRNAs and PIWI-like proteins in leukemia and lymphoma. By evaluating their efficacy, accuracy, and utility as biomarkers and therapeutic targets, this systematic review seeks to contribute insights essential for advancing personalized oncology strategies. Understanding the implications of piRNAs and PIWI-like proteins in hematologic malignancies not only enriches our knowledge of cancer biology but also holds promise for refining diagnostic approaches and developing targeted therapies.6–10 This review underscores the importance of integrating molecular insights into clinical decision-making and highlights avenues for further research aimed at improving patient outcomes in leukemia and lymphoma.
This study was conducted as a systematic review of the scientific literature in the field of Oncology and Hematology, aiming to explore an overview of the role of piRNAs and PIWI-like proteins as biomarkers and therapeutic targets in leukemia and lymphoma. Its development followed the guidelines of the PRISMA11, 12 statement for the proper conduct of systematic reviews. The elaboration process in its various phases will be detailed below.
Eligibility criteria
The eligibility of the studies was based on the fact to evaluate the role of piRNAs and PIWI-like proteins as biomarkers and therapeutic targets in leukemia and lymphoma.
Inclusion criteria
Exclusion criteria
Information sources and search strategy
The initial searches were conducted in July 2024 by combining the terms "Lymphoma", “Leukemia” and "piRNA" in PubMed and Sciencedirect databases. Subsequently, it was expanded with a combination, using boolean operators AND and OR as appropriate, of the terms “Leukemia”, “Lymphoma”, “piRNA”, “PIWI-Like proteins”. These searches yielded a considerable number of results.
The systematic search was conducted again on Pubmed and ScienceDirect, including all results found since information on this topic is scarce. The combination of terms that yielded the best results in both search engines was as follows: (Lymphoma OR Leukemia) AND (((piRNA) OR (piRNAs)) OR (PIWI-Like proteins)). Specifically, 26 results were obtained. Before proceeding with the article selection, inclusion and exclusion criteria were defined. Google Scholar was also used with various combinations of the mentioned search terms to verify if any articles that should have been included were overlooked and to confirm the inclusion of grey literature.
Selection process
The selection process was carried out in 3 phases: identification, screening and inclusion. The identification phase was carried out based on the search strategy and all the results were included, due to their limited number. In the screening phase, Rayyan13 was used, which is a web-based software for the initial reading of the selected studies using a semi-automated process. The review authors, anonymously and independently, read the title and abstract of each identified study and the inclusion and exclusion criteria were applied to classify each study as "Included", "Maybe" and "Excluded". Conflicts were resolved by 1 review author. Finally, for the inclusion phase, the selected articles underwent a full-text review by the review authors.
According to these criteria, using the Rayyan13 tool, the abstract of each article was read. Ultimately, 21 articles were excluded for not meticulously meeting the inclusion criteria (n=17) and for deviating from the objective theme (n=4). Finally, 5 articles met the inclusion criteria and were selected for the systematic review.
Data collection process, data items and quality assessment
Data extraction was carried out by all review authors. Any differences were resolved through discussions with the rest of the review authors until consensus was reached. A full-text review of each manually selected article was carried out with the help of Excel 2024 software to tabulate the information to be collected and Rayyan13. The data and variables sought in each study include: names of authors, year of publication, title, research design, objective, methodology, population (whether cell lines, animal models or humans) in relation to tumor type and results.
Risk of bias and quality assessment
The risk of bias was assessed by 1 independent author and any differences were discussed with a second author until consensus was reached. For in vivo studies, SYRCLE's14 tool was used. For in vitro studies, the CONSORT15 modified checklist for in vitro studies was used. For each study, an overall score of at least 50% was set as a cutoff value to be included. AMSTAR216 was used to assess the quality of this systematic review.
We searched in two databases (PubMed and ScienceDirect) for relevant literature and identified 26 papers. After removing duplicates (n = 0), we screened titles and abstracts (n = 26) and removed review articles and unrelated papers (n =21). Finally, 5 studies were included in this systematic review (Figure 1).
The actual research analyzed 5 articles that revealed the role of piRNAs and PIWI-like proteins as biomarkers and therapeutic targets in leukemia and lymphoma (Table 2).
|
Author |
Malignancy |
Summary |
|
Ding W |
ALL |
This study examined the role of PIWIL1 polymorphisms in pediatric acute lymphoblastic leukemia (ALL) relapse. In a case-control design with 785 cases and 1,323 controls, it found that the PIWIL1 SNP rs1106042 A>G increased ALL risk, while rs10773771 C>T decreased it. Rs1106042 GA/AA was linked to poor prognosis across various patient subgroups, and specific haplotypes (CAGT, TACC, TACT, TAGT) were associated with higher relapse susceptibility. Thus, rs1106042 A>G is a potential predictor of increased ALL risk and poor prognosis in eastern Chinese children. |
|
Bamezai S |
AML |
PIWIL4, an RNA-binding protein overexpressed in acute myeloid leukemia (AML) patients, is crucial for leukemic stem cell function and AML growth but is not necessary for healthy hematopoietic stem cells. In AML cells, PIWIL4 primarily interacts with mRNA linked to protein-coding regions and cancer-associated genes, rather than piwi-interacting RNA. Depleting PIWIL4 downregulates leukemia stem cell-associated genes, upregulates DNA damage signaling, and prevents R-loop accumulation, which maintains gene expression and prevents DNA damage in AML cells. This depletion also increases sensitivity to ATR pathway inhibition, presenting a potential therapeutic target in AML. |
|
Nasseri S |
AML |
Acute myeloid leukemia (AML) involves excessive myeloid cell production in the bone marrow. The piRNA hsa-piR-32877 is upregulated in AML and its downregulation via antisense LNA GapmeRs reduces myeloid cell proliferation and induces apoptosis. Experiments using human bone marrow blast cells and the M-07e cell line showed that antisense LNA GapmeRs effectively downregulate hsa-piR-32877, decrease cell proliferation, and increase apoptosis. This suggests that targeting hsa-piR-32877 could be a novel therapeutic approach for inhibiting leukemic cell proliferation in AML. |
|
Han H |
DLBCL |
Diffuse large B-cell lymphoma (DLBCL) progression is influenced by genetic and epigenetic changes. The role of N6-methyladenosine (m6A) in DLBCL is unclear, but high piRNA-30473 expression promotes an aggressive DLBCL phenotype. Depletion of piRNA-30473 reduces proliferation, induces cell cycle arrest, and decreases tumor growth in xenograft models. piRNA-30473 is linked to overall survival and enhances m6A levels by upregulating WTAP, an m6A methylase, which in turn increases hexokinase 2 (HK2) expression, promoting DLBCL progression. The piRNA-30473/WTAP/HK2 axis highlights the significance of m6A modification in DLBCL, aiding in prognostic and therapeutic development. |
|
Kaur G |
CLL |
Abnormal expression patterns of regulatory small non-coding RNAs (sncRNAs), such as miRNAs, piRNAs, and snoRNAs, are critical in cancer development. This study identified eight differentially expressed miRNAs in Chronic Lymphocytic Leukemia (CLL) via genome-wide small RNA sequencing. Three miRNAs (miR-1295a, miR-155, miR-4524a) were up-regulated, and five (miR-30a, miR-423, miR-486*, let-7e, miR-744) were down-regulated, validated by RQ-PCR. Additionally, seven novel sequences, including tRNA/piRNAs (piRNA-30799, piRNA-36225) and snoRNA (SNORD43), showed elevated expression in CLL. Multivariate analysis revealed miR-4524a and miR-744 as significant markers for risk and time to first treatment, suggesting their potential integration into CLL risk stratification and prognostication. |
Table 2 Characteristics of the reviewed studies
ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; DLBCL, diffuse large b-cell lymphoma; CLL, chronic lymphocytic leukemia
Role of piRNA in acute lymphoblastic leukemia
Ding W, Wang D, and colleagues conducted a comprehensive study to elucidate the role of PIWIL1 gene polymorphisms in pediatric acute lymphoblastic leukemia (ALL) relapse susceptibility among Chinese children. The study, involving 785 cases and 1,323 controls, employed a case-control design and multiple logistic regression to assess the risk associated with five single-nucleotide polymorphisms (SNPs) of the PIWIL1 gene. The results revealed that the rs1106042 A>G SNP increased the risk of ALL, while rs10773771 C>T decreased it. Stratified analyses indicated that rs1106042 GA/AA had a detrimental effect on various subgroups, including children under 120 months, those with high white blood cell counts, and several immunophenotypes and karyotypes. Conversely, rs10773771 TC/CC showed a protective effect in children with the TEL-AML fusion gene. Haplotype analysis further demonstrated that certain haplotypes were significantly associated with increased relapse susceptibility. The findings suggest that PIWIL1 rs1106042 A>G is linked to increased ALL risk and poor prognosis, while rs10773771 C>T is associated with reduced risk, providing insights into potential genetic markers for ALL prognosis and therapy.17
Role of piRNA in chronic lymphocytic leukemia
Kaur G and colleagues conducted genome-wide small RNA sequencing to identify differentially expressed miRNAs in Chronic Lymphocytic Leukemia (CLL). They found three upregulated (miR-1295a, miR-155, miR-4524a) and five downregulated (miR-30a, miR-423, miR-486*, let-7e, miR-744) miRNAs in CLL, validated by RQ-PCR. Additionally, seven novel sequences were identified, including tRNA/piRNAs (piRNA-30799, piRNA-36225) and snoRNA (SNORD43). Multivariate analysis revealed significant associations between miR-4524a, miR-744, and clinical outcomes, highlighting the potential of these miRNAs as biomarkers for CLL prognosis and treatment stratification.18
Role of piRNA in diffuse large b-cell lymphoma
Han H and colleagues studied the impact of piRNA-30473 on diffuse large B-cell lymphoma (DLBCL) and discovered that high expression of piRNA-30473 promotes tumorigenesis and poor prognosis by enhancing m6A RNA methylation. They showed that piRNA-30473 upregulates the m6A methylase WTAP, which in turn increases m6A levels on the target gene HK2, facilitating DLBCL progression. Inhibition of piRNA-30473 reduced tumor growth in xenograft models and improved overall survival, suggesting that the piRNA-30473/WTAP/HK2 axis could be a valuable prognostic marker and therapeutic target in DLBCL.19
Role of piRNA in acute myeloid leukemia
Bamezai S and colleagues explored the role of PIWIL4, an RNA-binding protein, in acute myeloid leukemia (AML). Their study found that PIWIL4, overexpressed in AML patients, is critical for leukemic stem cell function and AML progression but is dispensable for healthy hematopoietic stem cells. PIWIL4 primarily interacts with mRNA rather than piRNAs, affecting genes linked to cancer and myeloid progenitors. Depletion of PIWIL4 in AML cells led to downregulation of leukemia stem cell genes and upregulation of DNA damage signaling, highlighting PIWIL4's role in maintaining genomic integrity. Furthermore, PIWIL4 was shown to prevent DNA damage and replication stress, and its depletion increased sensitivity to ATR pathway inhibitors, suggesting PIWIL4 as a potential therapeutic target in AML.20
Nasseri S, Sharifi M, and Mehrzad V investigated the effects of suppressing hsa-piR-32877 in acute myeloid leukemia (AML) cells using antisense LNA GapmeRs. Their study demonstrated that hsa-piR-32877 is up-regulated in AML and that its downregulation led to decreased proliferation and increased apoptosis in both patient-derived bone marrow blast cells and the M-07e cell line. The findings indicate that targeting hsa-piR-32877 could serve as a novel therapeutic approach to inhibit leukemic cell proliferation in AML (Figure 2).21
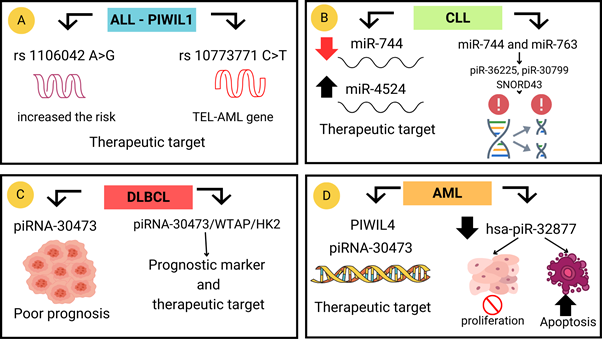
Figure 2 piRNA for diagnosis and treatment of leukemia and lymphoma. A) rs1106042 A>G and rs10773771 C>T are used as therapeutic targets.17 B) Induction of piRNA-30799, piRNA-36225 and SNORD43 generates mutations, while increased miR-4524 functions as a therapeutic target.18 C)Increased piRNA-30473 leads to tumorigenesis and poor prognosis, but its complex may function as a piRNA-30473/WTAP/HK2 therapeutic target.19 D) PIWIL4 protects against DNA damage and inhibition of hsa-piR-32877 generates apoptosis and decreases cell proliferation.20,21.
Our systematic review synthesizes current evidence regarding the involvement of PIWI-interacting RNAs (piRNAs) and PIWI proteins in hematologic malignancies, specifically acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), diffuse large B-cell lymphoma (DLBCL), and chronic lymphocytic leukemia (CLL). This comprehensive analysis underscores significant associations between genetic variants and disease susceptibility, as well as the functional roles of piRNAs in cancer pathogenesis.22–27
Firstly, our review highlights a study investigating PIWIL1 gene polymorphisms in pediatric ALL among Chinese children, revealing insights into genetic predisposition. Notably, variants rs1106042 A>G and rs10773771 C>T were identified as modulators of ALL risk, with rs1106042 A>G showing an increased susceptibility, particularly in younger patients and those with specific clinical profiles such as elevated white blood cell counts. Conversely, rs10773771 C>T demonstrated a protective effect, particularly in subtypes of ALL characterized by distinct genetic abnormalities. These findings underscore the intricate interplay between genetic variability and leukemia susceptibility, suggesting implications for personalized risk assessment and therapeutic strategies.28–33
Secondly, our synthesis of evidence regarding PIWIL4 in AML underscores its overexpression and pivotal role in leukemic stem cell maintenance and disease progression. PIWIL4, an RNA-binding protein involved in mRNA metabolism and genomic stability regulation, was found to be crucial in regulating pathways associated with leukemia stem cells and DNA damage response. Depletion studies demonstrated its potential as a therapeutic target in AML management. Additionally, research on hsa-piR-32877 in AML highlighted its aberrant expression and therapeutic promise. Suppression of hsa-piR-32877 using antisense LNA GapmeRs effectively reduced AML cell proliferation and induced apoptosis, suggesting a novel therapeutic avenue in AML treatment.34–36
Thirdly, our review delved into the oncogenic role of piRNA-30473 in DLBCL, elucidating its impact on tumor growth and prognosis. Elevated expression of piRNA-30473 correlated with aggressive DLBCL phenotypes and adverse clinical outcomes, mediated through its regulation of m6A RNA methylation and downstream oncogenic pathways. Targeting piRNA-30473 exhibited therapeutic efficacy in preclinical models, positioning it as a potential prognostic biomarker (marker to measure the effectiveness of the treatment) and therapeutic target in DLBCL.23,24,35
Lastly, our synthesis integrated findings on deregulated miRNAs in CLL, highlighting their pivotal roles in disease pathogenesis and clinical outcomes. Altered expression patterns of miRNAs, piRNAs, and snoRNAs in CLL underscored their potential as biomarkers for risk stratification and treatment response prediction. Notably, miR-4524a and miR-744 emerged as significant prognostic markers, correlating closely with clinical outcomes in CLL patients.1–3,32,36
This systematic review provides a comprehensive synthesis of the current literature on PIWI-interacting RNAs (piRNAs) and PIWI proteins in hematologic malignancies. The reviewed studies underscore significant associations between genetic variants of PIWI genes and susceptibility to acute lymphoblastic leukemia (ALL) and other hematologic cancers. Additionally, the functional roles of piRNAs in acute myeloid leukemia (AML), diffuse large B-cell lymphoma (DLBCL), and chronic lymphocytic leukemia (CLL) highlight their potential as biomarkers, therapeutic targets, and prognostic indicators.
The findings reveal that genetic polymorphisms in PIWI genes, such as PIWIL1, contribute to leukemia susceptibility, with specific variants influencing disease risk and clinical outcomes. Moreover, the oncogenic roles of PIWI proteins, including PIWIL4, in maintaining leukemic stem cell function and promoting disease progression underscore their therapeutic potential in targeted interventions.
Furthermore, the aberrant expression and functional implications of specific piRNAs, such as hsa-piR-32877 and piRNA-30473, in leukemia and lymphoma highlight their promise as novel therapeutic targets. The suppression of these piRNAs demonstrates efficacy in preclinical models, suggesting their translational potential in clinical settings. Overall, this review underscores the complex roles of piRNAs and PIWI proteins in hematologic malignancies, paving the way for future research aimed at elucidating their precise mechanisms of action and exploring innovative therapeutic strategies.
Future research directions should focus on several key areas to advance our understanding and clinical applications of piRNAs and PIWI proteins in hematologic cancers:
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Thanks to Bryan, my childhood friend, for showing us that cancer makes no exceptions. May he rest in peace.
Contributors played a substantial role in con-ception, design, acquisition, analysis, interpretation, writing, and critical review of the manuscript. All authors approved the final content and accept responsibility for its accuracy and integrity.
All authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers' bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent-licensing arrangements), or non-financial interest (such as personal or professional relationships, affiliations, knowledge or beliefs) in the subject matter or materials discussed in this manuscript.

©2024 Reyes, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.