eISSN: 2469-2778


Research Article Volume 8 Issue 6
Raisa Gorbacheva Memorial Research Institute for Pediatric Oncology, Hematology and Transplantation at the Pavlov First Saint Petersburg State Medical University, Russia
Correspondence: Nikolay N Mamaev, Raisa Gorbacheva Memorial Research Institute for Pediatric Oncology, Hematology and Transplantation at the Pavlov First Saint Petersburg State Medical University, 197022, Roentgena St., 12. St. Petersburg, Russian Federation, Russia
Received: December 05, 2020 | Published: December 31, 2020
Citation: Mamaev NN, Shakirova AI, Barkhatov IM et al. Crucial role of BAALC-expressing leukemic precursors in origin and development of posttransplant relapses in patients with acute myeloid leukemias. Hematol Transfus Int J. 2020;8(6):127?131. DOI: 10.15406/htij.2020.08.00240
acute myeloid leukemia, hematopoietic stem cell transplantation, posttransplant relapse, BAALC, WT1, combined overexpression
alloHSCT, allogeneic hematopoietic stem cell transplantation; AML, acute myeloid leukemia; PTR, posttransplant relapses, PCR, polymerase chain reaction; qRT-PCR, quantitative real time PCR;, BAALC, Brain and Acute Leukemia, Cytoplasmic gene; WT1, Wilm’s 1 tumor gene; FAB, French-American-British; M1-M7, FAB variants of AML; OS, overall survival; RFS, relapse-free-survival, CIR, cumulative incidence relapses
Despite the obvious advances in understanding of acute myeloid leukemia (AML) pathogenesis and successful treatment of some AML variants (e. g. M3), this problem has not been resolved even with the support of hematopoietic stem cell transplantation (HSCT). The main reason of HSCT failure is often posttransplant relapse (PTR) which can be provided by some leukemic precursors.1
The first step toward the elucidation of basic mechanisms of PTR origin was made by Italian investigators, who developed a method of highly sensitive quantitative evaluation of WT1-expressing blast elements burdens based on real-time PCR2,3 and successfully tested it at clinics.4,5 The second step has been done in this direction after opening and clinical investigation of Brain and Acute Leukemia, Cytoplasmic gene, e.g. BAALC. This gene was discovered to be expressed in the cytoplasm of neuroectoderm-derived tissues, especially in neurons6 as well as in bone marrow CD34+ cells.7 It is localized on the long arm of chromosome 8 and is overexpressed in 40-50% of AML patients.8,9 First evidence has recently been obtained that specific leukemic precursors with immunophenotype CD34+CD38- are responsible for the high BAALC mRNA production in patient’s bone marrow. 10 The number of such cells as well as the BAALC gene expression level were the highest in patients with M0, M1 and M2 AML FAB variants,11–13 but they were the lowest in M3 FAB variant,14,15 that may be explained by different types of leukemia initiating cells (LICs). The latter were discovered after elegant experiments with successful engrafting previously sorted leukemic precursors in immunodeficient mice16,17. As a result, at least two kind of LICs immature and mature were recognized. The first were more characteristic for M0, M1, M2 and M4 FAB variants of AML10, whereas the second one was responsible for hematopoiesis in APL 18. Meanwhile, immature LICs reveal immunophenotype CD34+/CD38- being capable to selective expression of pan-specific molecular marker BAALC10 . As for mature LICs, they are responsible for leukemic hematopoiesis in APL and along with blasts can express another pan-specific molecular marker WT1. First of all, this assumption is based on many cases of AML with higher levels of WT1 gene expression and lower number of blasts in tested samples of bone marrow as before, so after HSCT performing [Cilloni et al 2002, 2009; Mamaev 2019, 2020]. Secondly, it was recently shown that levels of WT1 expression in some patients with APL could be highest in AML group, whereas where as the number of blasts in the tested bone marrow samples could be lower 30 % [Mamaev et al, 2019; 2020]. Since levels of BAALC and WT1 expressions may be successfully determined by means of standard quantitative PCR in real time, a possibility appears to measure in such a way relative burden of BAALC-expression and less correctly (due to blasts) WT1–expressing relatively mature LICs.16–18
Developing this idea, we have recently showed the crucial role of BAALC-expressing leukemic precursors in origin and development of PTR19,22 The basic evidence was obtained on the group of AML patients (n=12), who had so called false remission just prior to conditioning and HSCT. As a result, BAALC gene overexpression was found in 3/12 (25%) patients in this group, that was associated with an increased incidence of PTR in all of them (p=0.002) as well as shortened event free survival (EFS) (p=0.019). When this molecular indicator of paired overexpression of BAALC and WT1 genes was tested in a larger AML patient cohort (n=110) with various cytological and cytogenetic variants, we revealed the important role of BAALC-producing precursors in origin and further development of PTR in patients with M1, M2, M4, and M5 FAB variants.18,19,22 Additionally, similar data have been obtained also in patients APL with PTR which will be discussed here in details too.
Our retrospective study presents the data of BAALC and WT1gene expression levels measured in parallel during serial sampling of bone marrow taken from 50 patients diagnosed with AML. All the patients were treated with alloHSCT at R. Gorbacheva Memorial Research Institute of Children Oncology, Hematology and Transplantation (St. Petersburg) from 2010 to 2016 years. The study group included 25 females and 25 males at the age of 1 to 60 years (median age of 25.8 years old). Among them were nine pediatric patients under the age of 17 year. Of notice, 7/50 included AML were EVI1-positive, although they did not contained in karyotypes any specific aberrations of loci 3q26. In all patients with different AML FAB-variants, serial BAALC and WT1 gene expression changes were evaluated individually in combination with counting of blast cell numbers in the same bone marrow samples. Written informed consent for all individuals was obtained in accordance with the Declaration of Helsinki.
Analysis of BAALC and WT1 gene expression levels
Total mRNA extraction from fresh bone marrow samples, its reverse transcription and quantitative estimation of the BAALC gene expression level by quantitative real-time PCR (qRT-PCR) were performed as previously described [20]. BAALC transcript copy numbers (CN) were determined using appropriate BAALC RQ-Kit (Inogene, Russia), including plasmid standards for constructing of calibration curves for the BAALC gene and reference gene ABL1. Basic control time points for bone marrow sampling were as follows: at diagnosis (e.g., D-80), just before alloHSCT, prior to the conditioning (D0) and after alloHSCT, i.e. on D+30, D+60, D+90, D+150 and later post-transplant. The bone marrow sampling was obligatory in cases of PTR occurrence. A median follow-up time after HSCT was 7 months (range from 0.6 to 52.5). The analysis of BAALC gene expression included 93 samples tested at clinical cytologically proven relapse, and 299 bone marrow aspirates that were taken during clinical remission. The relative BAALC gene expression level was calculated as a ratio of CNBAALC to CNABL1 and expressed in percents. The value of 31% was chosen as a common cut-off value to study clinical significance of BAALC gene overexpression before and after HSCT and to perform molecular monitoring of the interaction between leukemic precursors and blast cell populations. This value was higher than the maximum of BAALC gene expression level in patients with pre-transplant cytological remission, who did not show any clinical signs of the disease progression.
In parallel, the WT1 gene expression levels were determined in each sample at the same time points. The copy numbers of WT1 transcripts were determined by the same qRT-PCR method according to recommendations of European Leukemia Net group.4 The basal WT1gene expression level of 250 copies per 104 copies of ABL1 reference gene was applied to designate low and high WT1expressors.
Statistical analysis
Descriptive statistic methods for data with asymmetric distribution were used, with evaluation of the sample ranges and median values. Two-year relapse-free survival (RFS) and cumulative incidence of relapse (CIR) were calculated using Kaplan-Meier method, and the log-rank test was used to compare differences between survival curves. RFS and CIR were measured from D0 until the date of death, regardless of cause, or until terms of documented relapse, or last contact date. P < 0.05 was considered as statistically significant difference level. SPSS software version 22.0 (IBM corporation, Armonk, NY, USA) was used for statistical analysis.
The basic findings of our study are presented in Table 1. These data show higher expression BAALC gene closely related to increase burden of immature LICs to be presented in leukemic population of all tested FAB-cytological, cytogenetic and molecular variants of AML In most of them there was combined with higher expression of both tested genes indicated in the Table 1 by bold/ Therefore, many of them were transplanted at state of relapse which in turn could provide both shorter survival of these patients and death. It should be also noticed the combined BAALC and WT1 genes higher expression to be presented in cases of APL and AML with inv(16) which were characterized by aggressive clinical course (#25 and #40, respectively), whose detail clinical and laboratory details will be published additionally (Figure 1).
Patient, № |
FAB-variant |
Gender |
Age, years |
Cytogenetic group |
Number of HSCT |
Disease stage |
Post |
||||||||
Diagnosis |
D0 |
PTR |
|||||||||||||
BAALC, % |
WT1, CN |
Blasts, % |
BAALC, % |
WT1, CN |
Blasts, % |
BAALC, % |
WT1, CN |
Blasts, % |
|||||||
1 |
М1 |
m |
18 |
CK/del(7q) |
1 |
242 |
13542 |
93,2 |
n/d |
n/d |
n/d |
- |
- |
- |
730* |
2 |
М1 |
f |
19 |
t(8;21) + OA |
1 |
370 |
402 |
32 |
n/d |
n/d |
n/d |
- |
- |
- |
95 |
3 |
М1 |
m |
21 |
t(8;21) + OA |
1 |
n/d |
n/d |
n/d |
0,5 |
379 |
2.5 |
329 |
7858 |
70 |
306 |
4 |
М1 |
f |
25 |
NK |
1 |
n/d |
n/d |
n/d |
40 |
1764 |
13.2 |
- |
- |
- |
613 |
5 |
М1 |
m |
26 |
NK |
1 |
n/d |
n/d |
n/d |
2 |
47 |
2.6 |
2 |
35 |
48 |
179 |
6 |
М1 |
f |
26 |
CK, EVI1+ |
1 |
130 |
10493 |
84,2 |
n/d |
n/d |
n/d |
15 |
488 |
11,6 |
692 |
2 |
125 |
6500 |
51,2 |
n/d |
n/d |
n/d |
125 |
6500 |
51,2 |
||||||
154 |
2930 |
22 |
|||||||||||||
7 |
М1 |
f |
32 |
NK |
1 |
n/d |
n/d |
n/d |
21 |
40 |
2 |
37.1 |
102 |
7,8 |
672+ |
8 |
М1 |
f |
44 |
CK/del(7q), del(5q) |
1 |
n/d |
n/d |
n/d |
29 |
2355 |
8,8 |
n/d |
n/d |
n/d |
45 |
9 |
М1 |
f |
48 |
n/d |
1 |
17 |
386 |
60 |
n/d |
n/d |
n/d |
117 |
281 |
40 |
160? |
10 |
М1 |
f |
54 |
NK |
1 |
n/d |
n/d |
n/d |
32 |
18439 |
79,2 |
77 |
32981 |
35,4 |
140? |
11 |
M1 |
f |
60 |
NK |
1 |
2619 |
1728 |
96,6 |
3 |
29 |
0.6 |
378 |
197 |
37 |
382 |
12 |
М2 |
m |
8 |
t(8;21) + OA |
1 |
321 |
1236 |
69,6 |
22 |
7 |
4,2 |
- |
- |
- |
469+ |
13 |
М2 |
m |
9 |
NK |
1 |
99 |
10789 |
21,2 |
27 |
4 |
1,6 |
- |
- |
- |
730* |
14 |
М2 |
f |
15 |
t(8;21) + OA |
2 |
89 |
37 |
71,2 |
34 |
322 |
9 |
67 |
2033 |
45 |
730* |
133 |
9929 |
75,6 |
|||||||||||||
15 |
М2 |
m |
15 |
NK |
1 |
0,6 |
37 |
8,8 |
n/d |
n/d |
n/d |
- |
- |
- |
730* |
16 |
М2 |
m |
28 |
OA |
1 |
n/d |
n/d |
n/d |
n/d |
n/d |
n/d |
23 |
5518 |
24 |
162 |
17 |
М2 |
m |
30 |
t(8;21) + OA |
2 |
4 |
725 |
26,8 |
n/d |
n/d |
n/d |
153 |
2363 |
23 |
730* |
47 |
2649 |
28 |
|||||||||||||
18 |
М2 |
m |
35 |
NK |
1 |
n/d |
n/d |
n/d |
34 |
4238 |
5 |
- |
- |
- |
20 |
19 |
М2 |
f |
38 |
CK/del(7q) |
1 |
n/d |
n/d |
n/d |
787 |
8756 |
59,4 |
- |
- |
- |
407 |
20 |
М2 |
m |
39 |
NK, EVI1+ |
1 |
n/d |
n/d |
n/d |
n/d |
n/d |
n/d |
83 |
18872 |
26 |
350 |
21 |
М2 |
f |
49 |
CK/del (5q) |
1 |
n/d |
n/d |
n/d |
14 |
4 |
6,5 |
n/d |
n/d |
n/d |
214 |
22 |
М2 |
m |
58 |
t(8;21) + OA |
1 |
n/d |
n/d |
n/d |
366 |
26100 |
45,8 |
n/d |
n/d |
n/d |
763 |
23 |
M3 |
f |
17 |
t(15;17), EVI1+ |
1 |
n/d |
n/d |
n/d |
0.8 |
2957 |
1.8 |
3 |
32684 |
22.4 |
160 |
24 |
M3 |
m |
18 |
t(15;17) |
1 |
0.05 |
4375 |
33.6 |
0.2 |
421 |
10 |
- |
- |
- |
730* |
25 |
M3 |
f |
45 |
t(15;17) |
1 |
n/d |
n/d |
n/d |
n/d |
n/d |
n/d |
90.4 |
10693 |
60 |
730* |
26 |
М4 |
m |
5 |
t(8;21)/ del(7q) + OA |
1 |
95 |
999 |
65,8 |
n/d |
n/d |
n/d |
35 |
411 |
12,6 |
730* |
27 |
М4 |
m |
6 |
CK/del(5q) |
1 |
982,1 |
183 |
54,6 |
n/d |
n/d |
n/d |
7118 |
296 |
41 |
779 |
28 |
М4 |
f |
16 |
NK |
1 |
34 |
1479 |
22,5 |
n/d |
n/d |
n/d |
- |
- |
- |
128 |
29 |
М4 |
m |
17 |
NK |
1 |
n/d |
n/d |
n/d |
5 |
51 |
3 |
6 |
362 |
7 |
197 |
30 |
М4 |
m |
17 |
NK |
1 |
n/d |
n/d |
n/d |
2 |
2258 |
12 |
- |
- |
- |
730* |
31 |
М4 |
f |
17 |
OA |
2 |
52 |
8043 |
76 |
41 |
3078 |
0,6 |
61 |
7809 |
51 |
454 |
61 |
7809 |
51 |
31 |
8407 |
2.4 |
69 |
2424 |
50,8 |
|||||||
32 |
М4 |
m |
18 |
NK |
1 |
n/d |
n/d |
n/d |
10 |
836 |
7,2 |
- |
- |
- |
61 |
33 |
М4 |
m |
19 |
NK |
1 |
27 |
14929 |
95 |
9 |
1319 |
25.4 |
- |
- |
- |
311 |
34 |
М4 |
f |
19 |
inv(16) + OA |
1 |
n/d |
n/d |
n/d |
25 |
96 |
10.6 |
- |
- |
- |
370* |
35 |
М4 |
f |
21 |
CK/del(7) EVI1+ |
2 |
107 |
867 |
17 |
n/d |
n/d |
n/d |
389 |
529 |
12 |
393 |
400 |
26036 |
24 |
|||||||||||||
36 |
М4 |
m |
21 |
NK |
1 |
n/d |
n/d |
n/d |
39 |
41 |
5,4 |
- |
- |
- |
518+ |
37 |
М4 |
f |
25 |
NK |
1 |
107 |
1790 |
82 |
n/d |
n/d |
n/d |
- |
- |
- |
730* |
38 |
М4 |
m |
27 |
inv(16) |
1 |
n/d |
n/d |
n/d |
20 |
1696 |
10 |
- |
- |
- |
459 |
39 |
М4 |
f |
28 |
OA |
1 |
n/d |
n/d |
n/d |
34 |
7367 |
7,6 |
n/d |
n/d |
n/d |
730* |
40 |
М4 |
f |
34 |
inv(16) |
1 |
n/d |
n/d |
n/d |
549 |
10819 |
11,6 |
0,5 |
7223 |
10.4 |
560 |
41 |
М4 |
m |
39 |
NK |
1 |
n/d |
n/d |
n/d |
503 |
3548 |
62 |
- |
- |
- |
614 |
42 |
М4 |
f |
45 |
NK |
1 |
n/d |
n/d |
n/d |
n/d |
13095 |
48,8 |
31 |
3381 |
n/d |
700? |
43 |
М4 |
f |
55 |
NK |
1 |
4 |
11686 |
68 |
n/d |
n/d |
n/d |
0,06 |
1020 |
20 |
101 |
44 |
М5 |
m |
11 |
del(7) +OA |
1 |
45 |
11753 |
88 |
n/d |
n/d |
n/d |
- |
- |
- |
730* |
45 |
М5 |
m |
22 |
t(9;11) |
1 |
17 |
25 |
94,4 |
0,6 |
6 |
1,4 |
10 |
25 |
9,6 |
730* |
46 |
М5 |
f |
32 |
CK/del(7q) |
1 |
n/d |
n/d |
n/d |
272 |
3779 |
31,6 |
n/d |
n/d |
n/d |
142 |
47 |
М5 |
m |
37 |
CK/del(7), EVI1+ |
1 |
2 |
177 |
8 |
2 |
77 |
3,8 |
24 |
4542 |
21 |
423 |
48 |
М5 |
m |
55 |
NK |
1 |
1,3 |
9631 |
40 |
n/d |
n/d |
n/d |
- |
- |
- |
730* |
49 |
М7 |
f |
1 |
CK, EVI1+ (chromothripsis) |
1 |
n/d |
n/d |
n/d |
n/d |
n/d |
n/d |
9 |
3315 |
60 |
148 |
50 |
М7 |
f |
3 |
EVI1+ |
1 |
0.25 |
282 |
12.8 |
3 |
3693 |
13.4 |
0.3 |
1049 |
20.8 |
258 |
Table 1 BAALC and WT1 gene expression levels and other clinical and hematological characteristics of patients at different stages before and after alloHSCT (only samples with maximal blast cells burden are pointed).
Conventional symbols: PTR, posttransplant relapse; m, male; f, female; CN, copy number per 104copies of ABL1; CK, complex karyotype (>3 cytogenetic aberrations per metaphase); NK, normal karyotype; OA, other cytogenetic aberrations (<2); - – no PTR during two years after alloHSCT; n/d, no data; relapse. Bone marrow samples with combined BAALC and WT1 gene overexpression are shown in bold; * - censored data; ? – OS in these cases was incorrect due to absent of their correct death date.
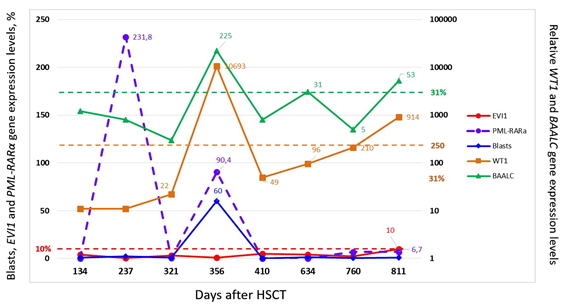
Figure 1 The results of serial measurements of BAALC. WT1? EVI1 and PML/RARa expression along to number blasts counting in the tested bone marrow samples from treated by HSCT APL patient (#25) at the stage of posttransplant relapse which evidence a participation in relapse formation of BAALC-expressing immature LICs.
The data presented here show for the first time the crucial role of BAALC gene overexpression at pre-conditioning and posttransplant stages in the origin and further development of post-transplant relapses. According to our findings even single BAALC gene overexpression at the day D0 as well as at PTR is associated with an unfavorable prognosis. The latter was especially bad in the cases with combined overexpression of BAALC and WT1 genes in the studied patients with different cytological and cytogenetic AML variants. In the cohort of patients involved in this investigation, such phenomenon was represented 33 times, including 13 of them at diagnosis, 8 on the day D0 and 12 at PTR. Importantly for us, that overexpression of gene BALLC was provided by immature CD34+CD38- precursors only,10,11 which allows recognize subtle changes of BAALC-expressing burdens without great problems.
. As for WT1 gene overexpression concerns this mechanism is not elucidated yet. In our opinion, another issue of WT1 expression along with traditionally appreciated blasts must be presented. Indirect evidence of it is seen in parallel measurements of BAAALC and WT1 along with blast number counting in the testes bone marrow samples from patients with APL. As known, the levels of WT1 expression in APL are highest among other subtypes of AML24 whereas APL cases with aggressive course reveal higher BAALC expression.25 Since, some of APL patents with higher WT1 expression can reveal relatively low number of blasts, it allows to draw assumption concerning another issue of WT1 expression in these APL patients than blasts. In our opinion, the main candidate for this role may be recently opened in APL by Patel et al25 so-called mature subtype of LIC which retains to be evidenced in direct sorting experiments. Meanwhile, BAALC/WT1 molecular platform which was tested by us, will open possibilities to obtain important data concerning mechanisms of treatment resistance and PTR formation in treated with HSCT AML patients on top precursor level.
So, the study hematopoiesis in patients with AML by means of molecular platform BAALC/WT1 allows to elucidate some subtle molecular mechanisms which could be responsible for formation both treatment resistance and aggressive course of AML. Since BAALC gene levels expression are closely associated with burden of its producing LICs it is possible to study different aspects of pathogenesis in various cytological, cytogenetic and molecular variants of AML. by means standard enough cheap and not time-consuming qRT-PCR
Authors declare no conflict of interests for this article.
NM, AS: concept and design
AS, IB, YG, TG, VK, ZR, MK, OP: submission of study materials
NM, AS: analysis and interpretation of data, drafting the manuscript
LZ: administrative support
All authors approved the final version of the manuscript.
We express gratitude to the transplantation team CIC 725 of Raisa Gorbacheva Memorial Research Institute for Pediatric Oncology, Hematology and Transplantation at the Pavlov First Saint Petersburg State Medical University and especially to the Laboratory of molecular hematology and transplantation.

©2020 Mamaev, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.