eISSN: 2576-4462


Research Article Volume 3 Issue 4
Medicinal Plants Research Center, Department of Cell and Molecular Biology, Universiti Putra Malaysia, Malaysia
Correspondence: Daryush Talei, Medicinal Plants Research Center, Shahed University, Shahed University, Department of Cell and Molecular Biology, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, 43400 UPM, Serdang, Selangor DE, Malaysia
Received: May 09, 2019 | Published: July 12, 2019
Citation: Talei D. Morpho-physiological diversity of Andrographis paniculata accessions using multivariate analysis. Horticult Int J. 2019;3(4):166-170. DOI: 10.15406/hij.2019.03.00126
Andrographis paniculata is a medicinal herb in the family Acanthaceae. Knowledge of Morpho-physiological diversity improves the efficiency of germplasm conservation and development. In order to evaluate Morpho-physiological diversity of 32 accessions of A. paniculata from different states of Malaysia, a Randomized Complete Block Design with 32 different accessions and three replicates was carried out. The results showed highly significant differences among the accessions in terms of most of the studied traits, while there were no significant differences among the accessions in terms of number of leaf and root fresh weight. Variance analysis based on seven states showed that there were no significant differences among the accessions in terms of most of the studied Morpho-physiological traits inside the each state. The highest total dry weight (TDW) (0.41 g) was belonged to accession No. 11340 from Kelantan, while the lowest TDW (0.12 g) was belonged to accession No. 11314 from Terengganu. The correlation between most of the studied traits was high significant and positive. The cluster analysis based on the studied traits of 32 accessions produced three groups. The first group comprised of 5 accessions, the second group consisted of 10 accessions and the third group contains 17 accessions. Overall, the outcomes of the present study were indicated the presence of high genetic variability among the A. paniculata accessions. Our findings suggest that the plants belong to different clusters can be used for hybridization to generate useful recombinants in the segregating generations, the genetics and breeding programs for improvement of A. paniculata.
Keywords: Andrographis paniculata, cluster analysis, genetic diversity, morphological trait, PCA analysis.
Andrographis paniculata is a medicinal herb in the family Acanthaceae that rich in active compounds of therapeutic importance. The plant extract contains three major compounds namely diterpenes, flavonoids and stigmasterols1 with the active compounds identified as andrographolide (ANDRO), neoandrographolide (NAG) and 14-deoxy-11, 12-didehydroandrographolide (DDAG).2 The herb exhibited a wide scope of pharmaceutical properties such as anti-HIV,3 anti-H1N1,4 anticancer,5 anti-hepatitis,6 anti-flammatory7 and anti-diarrhea.8 These compounds are produced mainly from aerial organs of the plant especially leaves.
One of the important requirements for crop planning in order to achieve high yield and desirable quality, especially in the case of medicinal plants, is the evaluation of genetic variation of plants. In breeding programs, genetic diversity is the first step that has been created over the years and is one of the most important factors in the survival of creatures, including plants, for adaptation to environmental changes. Genetic diversity can be estimated based on genealogy analyzes using various methods including morphological traits, protein markers, and molecular markers.9 The objective of this study was to evaluate morphological and agronomical diversity among 32 accessions of Andrographis paniculata collected from seven different states of Malaysia and to estimate genetic relationships among the accessions which can be utilized for breeding program and phytochemical study.
Plant materials
A total of thirty-two accessions of A. paniculata seeds were used in this study. The seeds were kindly provided by Agro Gene Bank, Universiti Putra Malaysia, Serdang, Selangor, Malaysia (Table 1).
Acc. No |
State |
Acc. No |
State |
Acc. No |
State |
Acc. No |
State |
11304 |
Johor |
11349 |
Kelantan |
11249 |
N.Sembilan |
11266 |
Perak |
11306 |
Johor |
11204 |
N.Sembilan |
11301 |
Pahang |
11159 |
Selangor |
11325 |
Johor |
11216 |
N.Sembilan |
11313 |
Pahang |
11176 |
Selangor |
11352 |
Johor |
11218 |
N.Sembilan |
11328 |
Pahang |
11179 |
Selangor |
11303 |
Kelantan |
11228 |
N.Sembilan |
11345 |
Pahang |
11314 |
Terengganu |
11329 |
Kelantan |
11231 |
N.Sembilan |
11347 |
Pahang |
11317 |
Terengganu |
11339 |
Kelantan |
11238 |
N.Sembilan |
11264 |
Perak |
11323 |
Terengganu |
11340 |
Kelantan |
11245 |
N.Sembilan |
11265 |
Perak |
11348 |
Terengganu |
Table 1 Accessions number and location of 32 accessions from different states of Malaysia
Seed germination and growth conditions
The seeds were scarified with sand Paper11 and were then surface sterilized with 10% sodium hypochlorite (NaClO) solution for 10 minutes12 and thoroughly rinsed with distilled water. The seeds were then sown in Petri dishes containing No 2 filter paper moistened with a uniform volume of sterile water. The Petri dishes were sealed with parafilm to prevent any water loss and then incubated in a growth chamber at controlled temperature, light and humidity (light/dark regime of 14/10h at 28-30oC, relative humidity 60- 75%). The seedlings at two-leaf stage were transferred into the Jiffy medium. The seedlings 30 days after being cultured in the Jiffy media were transferred into pots containing soil, Pit mousse, sand based on randomized complete block design with 32 accessions of A. paniculata and three replicates. Each plant was irrigated every two days with Hoagland solution. Before flowering stage all plants were harvested and the data on morphological and physiological traits were measured. The dry shoot and root weights (SDW and RDW) after drying at the 68 oC for 48 hours were measured.
Determination of chlorophyll and protein contentThe chlorophyll content of leaves was measured using Chlorophyll Meter XT SPAD-502 equipment. To determination the protein content, one gram fresh leaf from the 32 different A. paniculata accessions was grounded in liquid nitrogen using pre-cooled mortar and pestle to obtain a fine powder and then homogenized with 2mL of the HEPES/KOH buffers according to the method of Talei et al.13 Finally, the total protein concentration was determined by the Bradford method (1976) at 595 nm, using a spectrophotometer (Lambda 25, UV/VIS).
Statistical analysisThe SPSS software version 24 was used for variance analyses and Duncan’s multiple range test at significance level of P≤0.01 and the cluster analysis was performed using Unweighted Pair Group Method with Arithmetic Average (UPGMA) and PCA analysis using JMP software version 10.
The results of analysis of variance showed that there is a significant difference between the accessions based on most of the studied traits (P≤0.01), while there was no significant difference between the accessions based on leaf number, average leaf width, fresh weight root, chlorophyll and protein content (Table 2). The results of variance analysis also showed that there is no significant difference between different accessions within each state based on most morphological and physiological traits, while there were high significant differences between different accessions between states based on most morphological and agronomic traits. The highest dry weight (0.41±0.03 g) belonged to the 11340 accession from the Kelantan state, while the lowest dry weight (0.12±0.01 g) belonged to the accession of 11314 from the Terengganu state. The results indicated that the highest plant height (13.0±0.76 cm) belonged to the 11340 accession from the Kelantan state, while the lowest plant height (4.83±1.09 cm) belonged to the accession of 11314 from the Terengganu state. The results also showed that the highest number of leaf (15.7±0.03 g) belonged to the 11306 accession from the N.Sembilan state, while the lowest number of leaf (9±0.58 g) belonged to the accession of 11314 from the Terengganu state. There were no significant differences among different accessions in terms of chlorophyll and protein contents, nevertheless the highest chlorophyll content (42.13±3.15 µg.g-1 FW) belonged to the accession of 11159 from the Selangor state, while the lowest chlorophyll content (33.40±2.34 µg.g-1 FW) was obtained. Protein variation due to different accessions was between 30.57 to 26.29 µg.g-1 FW.
Source of variance |
df |
Mean square |
|||||||||||
NL |
PH |
MLL |
MWL |
SFW |
SDW |
RL |
RFW |
RDW |
TDW |
Chl |
Protein |
||
Accession |
31 |
9.98ns |
10.30* |
5.92* |
0.64ns |
0.99** |
0.019** |
3.98* |
0.02ns |
0.000** |
0.023** |
12.90ns |
3.38ns |
Error |
62 |
6.98 |
6.43 |
3.19 |
0.43 |
0.52 |
0.003 |
2.48 |
0.01 |
0.000 |
0.004 |
8.74 |
3.58 |
Table 2 Variance analysis of 32 accessions of A. paniculata from different states of Malaysia in terms of morphological and physiological traits
NL, number of leave; PH, plant height (cm); MLL, average leave length; MWL, average width leave; SFW, shoot fresh weight (g); SDW, shoot dry weight (g); RL, root length (cm); RFW, root fresh weight (g); RDW, root dry weight (g); TDW, total dry weight (g); Chl, chlorophyll content
The correlation between the studied traits was very significant and positive, while the correlation between protein and other studied traits was no significant. The highest correlation was found between total dry weight and shoot dry weight (r=0.99), and the lowest correlation between root length and root dry weight (r=0.31) (Figure 1) (Table 3).
NL |
PH |
MLL |
MWL |
SFW |
SDW |
RL |
RFW |
RDW |
TDW |
Chl |
Protein |
|
NL |
1 |
|||||||||||
PH |
.784** |
1 |
||||||||||
MLL |
.774** |
.902** |
1 |
|||||||||
MWL |
.756** |
.896** |
.964** |
1 |
||||||||
SFW |
.886** |
.896** |
.919** |
.888** |
1 |
|||||||
SDW |
.595** |
.602** |
.614** |
.606** |
.676** |
1 |
||||||
RL |
.484** |
.580** |
.631** |
.616** |
.579** |
.359** |
1 |
|||||
RFW |
.619** |
.663** |
.590** |
.556** |
.736** |
.525** |
.497** |
1 |
||||
RDW |
.589** |
.541** |
.543** |
.543** |
.664** |
.789** |
.313** |
.696** |
1 |
|||
TDW |
.606** |
.608** |
.619** |
.612** |
.689** |
.998** |
.362** |
.554** |
.828** |
1 |
||
Chl |
.541** |
.519** |
.512** |
.530** |
.551** |
.426** |
.369** |
.399** |
.494** |
.442** |
1 |
|
Protein |
.089 |
.148 |
.045 |
.039 |
.084 |
.208* |
-.044 |
.212* |
.108 |
.201* |
-.120 |
1 |
Table 3 Correlation coefficient (r) among measured traits of A. paniculata from different states of Malaysia
NL, number of leave; PH, plant height; MLL, average leave length; MWL, average width leave; SFW, shoot fresh weight; SDW, shoot dry weight; RL, root length; RFW, root fresh weight; RDW, root dry weight; TDW, total dry weight; Chl, chlorophyll content
Cluster analysis is performed to classify the genotypes into homogenous groups based on the similarity/dissimilarity percentage between them. The UPGMA cluster analysis of the 32 A. paniculata accessions using the Ward method under salinity conditions based on morphological and physiological traits produced five clusters. Cluster 1 (red colour) contained ten accessions, cluster 2 (green colour) contained seven accessions, cluster 3 (blue colour) including six accessions, cluster 4 (brown colour) contained six accessions and cluster five (light green colour) including three accessions (Figure 1). The distance between leader and joiner accessions in cluster analysis based on all measured traits was 13.90. Clustering pattern confirmed that the genetic diversity is independent of a geographical area as accessions collected from different regions were accumulated in same clusters and those from the same region were also distributed into different clusters randomly.
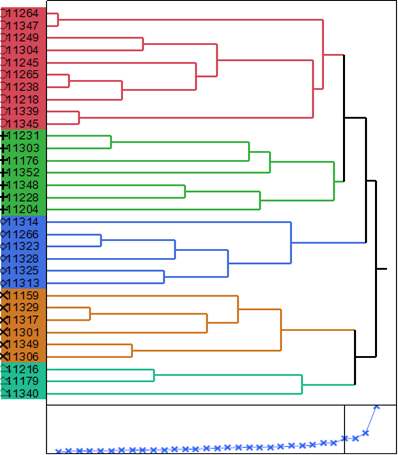
Figure 1 A dendrogram generated by UPGMA clustering method of 32 accessions of A. paniculata based on all measured traits.
The UPGMA cluster analysis of the seven states using the Ward method under salinity conditions based on morphological and physiological traits produced three clusters. Cluster 1 (red colour) contained two states (Selangor and Kelantan), cluster 2 (green colour) contained three states (N. Sembilan, Pahang and Johor) and cluster 3 (blue colour) including two states (Perak and Terengganu) (Figure 2). The distance between leader and joiner states in cluster analysis based on all measured traits was 7.32. In general, the results of the study showed that there is a high genetic diversity between the A. paniculata accessions in different states. The findings of research suggest that plants belonging to different clusters can be used in the hybridization program to produce recombinant genotyping, genetic diversity, and reproductive breeding programs for A. paniculata accessions.
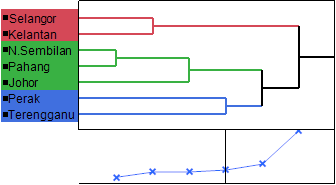
Figure 2 A dendrogram generated by UPGMA clustering method of seven states based on all measured traits.
Principal Component Analysis (PCA) is one of the multivariate analyses which reduces the more number of correlated variables to limited number uncorrelated individual variables known as Principal Components (PC), thereby, simplifies the multivariate data set. Therefore, for a better understanding of the effectiveness of the studied traits, principal components analysis (PCA) was performed using 12 characteristics on the scree plot (Figure 3). The PCA revealed two principal components (PC) with Eigen value>1 ranging from 1.6 to 8.78, which made up 81.71% of the total data variance. The PCA also revealed that all studied traits, except P and micro nutrients were located in the first component. The most important character in PC1 was total dry weight, while Mg2+ content was the least effective. Component matrices showed that physiological, phytochemical and biochemical traits contributed more effectively in other components. Nevertheless, since PC1 and PC2 represented 25.25% and 17.43% of the total variance respectively, so, the importance of the mentioned traits in PC1 and PC2 should be further looked into in future studies.
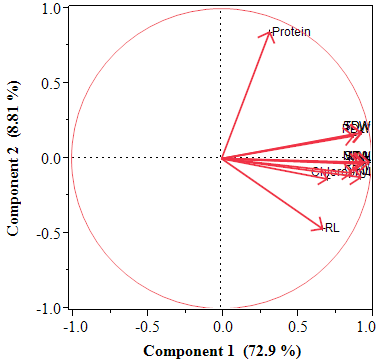
Figure 3 The principal components analysis (PCA) of 12 studied traits in 32 accessions of A. paniculata. The PCA revealed two principal components (PC).
To clarify the PCA results, the contribution by the studied traits was illustrated in a dendrogram. The morphological traits as well as the physiological traits were approximately segregated into different clusters. As an additional result, to simplify the variance analysis and to reduce the number of variables, one of the traits positioned in the same cluster could be omitted. The studied morphological and agronomical were clearly differentiated from biochemical traits grouped into separate clusters.
Broad-sense heritabilityBroad-sense heritability calculated for all characters revealed that among morphological and physiological characters, the h2 of shoot dry weight was the highest (0.86) and protein was the lowest (0.49) (Table 4). Unexpectedly, the heritability of salt tolerance have been only reported in very few traits such as shoot or root growth14–16 have reported the higher heritability of shoot and root length in Gossypium hirsutum. The present results confirm high heritability of shoot and root dry weight and total dry weight among agronomic traits as well as plant height and number of leave. Therefore, the underlying genetic mechanisms can be considered as direct criterion for assessing genetic diversity.
Components |
Characters |
|||||||||||
NL |
PH |
MLL |
MWL |
SFW |
SDW |
RL |
RFW |
RDW |
TDW |
Chl |
Protein |
|
9.98 |
10.30 |
5.92 |
0.64 |
0.99 |
0.02 |
3.98 |
0.02 |
0.00 |
0.02 |
12.90 |
3.38 |
|
16.96 |
16.73 |
9.11 |
1.07 |
1.51 |
0.02 |
6.46 |
0.03 |
0.00 |
0.03 |
21.64 |
6.96 |
|
0.59 |
0.62 |
0.65 |
0.60 |
0.66 |
0.86 |
0.62 |
0.67 |
0.80 |
0.85 |
0.60 |
0.49 |
|
Table 4 Components of variance and broad-sense heritability of morphological and agronomic characters in 32 A. paniculata accessions
NL, number of leave; PH, plant height; MLL, average leave length; MWL, average width leave; SFW, shoot fresh weight; SDW, shoot dry weight; RL, root length; RFW, root fresh weight; RDW, root dry weight; TDW, total dry weight; Chl, chlorophyll content; , genetic variance,, phenotypic variance and , broad-sense heritability
In general, the results of the study revealed that there is a high genetic diversity between the A. paniculata accessions in different states. The findings of research suggest that plants belonging to different clusters can be used in the hybridization program to produce recombinant genotyping, genetic diversity, and reproductive breeding programs for A. paniculata accessions.
None.
Conflict of interest Authors declares there is no conflict of interest.

©2019 Talei. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.