eISSN: 2576-4462


Research Article Volume 4 Issue 4
1Faculty of Agricultural Sciences, National University of Catamarca. Avda. Belgrano and Maestro Quiroga, Argentina
2Faculty of Agrarian Sciences, National University of the Littoral, Argentina
3National Council for Scientific and Technical Research, Argentina
4Faculty of Agronomy and Zootechnics, National University of Tucumán, Argentina
Correspondence: Di Barbaro Gabriela, Faculty of Agricultural Sciences, National University of Catamarca. Avda. Belgrano and Maestro Quiroga, Argentina
Received: July 01, 2020 | Published: August 4, 2020
Citation: Gabriela DB, Eleodoro DV, Celia BDW. Isolation of atmospheric nitrogen-free fixing bacteria from the endorhizosphere of Helianthus tuberosus and Smallanthus sonchifolius. Horticult Int J. 2020;4(4):129-134. DOI: 10.15406/hij.2020.04.00171
The objective of the work was to determine the presence of fixing bacteria free of atmospheric nitrogen in the endorhizosphere of topinambur (Helianthus tuberosus) and yacón (Smallanthus sonchifolius), cultivated in the Central Valley of the Province of Catamarca. Roots of each species were collected and the technique proposed by Döbereiner et al.1 for the isolation of N-free endorrhizospheric fixing bacteriatwo, Azospirillum sp. The isolates obtained were morphologically and physiologically characterized, and three different morphotypes of each plant species were selected for molecular identification. Eleven autochthonous fixative microorganisms free of atmospheric nitrogen present in the endorhizosphere of the crops under study were described, 6 in topinambur and 5 in yacón. All isolates belonging to the genus Pseudomonas. In topinambur, Pseudomonas sihuiensis and in yacón of Pseudomonas alcaligenes, P. resinovorans and P. sihuiensis. The isolation of autochthonous nitrogen-free bacteria native to P. sihuiensis, P. alcaligenes and P. resinovorans is reported for the first time in the endorhizosphere of topinambur (H. tuberosus) and yacón (S. sonchifolius), cultivated in soils of the Central Valley of the Province of Catamarca.
Keywords: jerusalem artichoke, yacón, catamarca, diazotrophs, pseudomonas
Molecular nitrogen (N2) that exists in the atmosphere is not easily assimilated by plants because the triple bond that unites the atoms that make up the molecule is difficult to break; plants then take advantage of atmospheric nitrogen through the metabolic process known as Biological Nitrogen Fixation (FBN).2 This process is carried out by certain soil microorganisms known as "nitrogen-fixing microorganisms" that convert nitrogen (Nt2) in ammonia (NH3) through the enzymatic complex called nitrogenase.3 Nitrogen-fixing microorganisms include symbiotic nitrogen-fixing fixers in association with plants, and free-living microorganisms that provide nitrogenous compounds for use by plants.4
Biological nitrogen fixation is the main source of this element to natural ecosystems and to agro-ecosystems without chemical fertilization,2,5,6 where this process contributes approximately 140 million tons of nitrogen per year, becoming the main entry of this element into the biosphere.3 By fixing nitrogen, these microorganisms influence the productivity of the crops and part of it is available in the soil as a carbon source for its microflora.2,6Among the genera of free-living FBN bacteria are Azomonas, Azospirillum, Azotobacter, Beijerinckia, Burkholderia, Clostridium, Derxia, Desulfovibrio, Enterobacter, Herbaspirillum, Klebsiella, Paenibacillus and Pseudomonas.7 However, the main free-fixing N-fixing microorganisms constituting the main biofertilizers correspond to the genera: Achromobacter, Acetobacter, Alcaligenes, Arthrobacter, Azotobacter, Cyanobacteria, Azospirillum and Pseudomonas.6,7
Due to their ability to fix nitrogen, nitrogen fixing bacteria are considered as candidates for the production of biofertilizers, which are manufactured in various countries of the world. Currently, biofertilizers are considered an important biotechnological tool, which consists in applying microorganisms that They help improve the availability of nutrients in the soil and thus reduce the effects caused by excess fertilizers. The use of biofertilizers in different crops has shown positive effects on soil fertility.6
Topinambur (H. tuberosus) and yacón (S. sonchifolius) crops date back to pre-Columbian times and have long been restricted to very small areas as they are adapted to ecological conditions in the Andes, where their use is strongly linked to the traditions of the peoples,8 and in recent years has gained great interest for its nutraceutical properties.
The consumption of foods with physiological and biochemical benefits are increasing, which is why the production of topinambur and yacón has generated interest in the last decade. Topinambur and yacón are perennial herbaceous plants that have sweet-tasting tuberous roots and are native to South America, where it is used in human food and in folk medicine for the treatment of diabetes, digestive problems, constipation and other diseases.9,10 These tubers contain natural antioxidants, fructans and fructooligosaccharides of interest to the nutraceutical industry.10,11–13
The H. tuberosus crop has a wide worldwide distribution. It is cultivated in various countries of the world, such as Italy, Australia,14 Germany,15 Austria, the Czech Republic,16 Denmark,17 Poland,18 Serbia,19 China,20 South Korea,21,22 Japan,23 Thailand,24 the United States,25 Cuba26 and Brazil.27 However, in the country there are references to some dispersed producers in the provinces of Buenos Aires, Córdoba, Entre Ríos, Mendoza, Río Negro, Chubut and San Luis.28
While, S. sonchifolius is currently distributed throughout a large part of the Andean territory from northern Ecuador to northwestern Argentina29,30 and has been successfully cultivated in several regions, including: Brazil, Czech Republic, China, Korea, Japan, New Zealand, Russia, Taiwan and the United States.9,31–33 In Argentina, yacón is only cultivated in the provinces from Jujuy, Salta and Tucumán.34,35 In Catamarca, both topinambur and yacón are non-traditional crops in the province and have been produced by a single producer in the area.
The objective of the work was to determine the presence of atmospheric N-free fixing bacteria in the endorhizosphere of topinambur and yacón plants in the province of Catamarca.
Specimens of topinambur (H. tuberosus) and yacón (S. sonchifolius) were collected from different bioassays that were carried out in the Central Valley of the Province of Catamarca at (28º28´07´´LS 65º46´60´´LO), place which is immersed in the semi-arid region of the northwest of the Argentine Republic.
Isolations of atmospheric N-free fixing bacteria from the endorhizosphere of topinambur and yacón plants were performed according to the procedure described by Döbereiner et al.1 in which the semi-solid and RC NFb culture media were used.1,36 For the identification of the isolated bacteria, the morphological, physiological and biochemical characters were established and examined according to the methods described by Holt et al.37 and Buchanan et al.38 For each isolate, the color of the colonies was determined in specific culture media (RC); morphology of the observed colony; response to Gram Nicolle and Shaffer and Fulton staining; acid/alkali production (semisolid NFb with bromothymol blue as indicator) and assimilation tests using different carbon sources (glucose, lactose, malic acid and starch). The tests of each bacterial isolate were carried out in triplicate (Figure 1).
For molecular identification, the isolates were sent to the Macrogen company (Korea), who were responsible for the extraction, amplification and sequencing of the DNA of each strain, and the bioinformatic analysis, which consisted of the analysis of the sequences obtained in the search for similar sequences in the GenBank through the BLAST program.
Eleven bacterial isolates were obtained from all the root samples studied. Six of them came from the endorhizosphere of topinambur and five from that of yacón. Based on the different morphotypes obtained, 3 topinambur and 3 yacon isolates were sent to Macrogen (Korea) for molecular identification. Morphologically, most of the isolates presented small and mucoid colonies, and corresponded entirely to non-sporulated Gram negative bacilli (Table 1).
Crop |
Isolated |
Characterization colony morphology |
Characterization |
16S rRNA gene% identity |
Topinambur |
Topi1 |
Red microcolonies and creamy in RC medium. Bacilli G (-), no esporulated |
Aerobic, mobile, glucose (-), lactose (-). |
Pseudomonas sihuiensis |
Topi1A |
Yellowish microcolonies and mucosa in RC medium. Bacilli G (-), no esporulated |
Aerobic, mobile, glucose (+), lactose (-). |
Pseudomonas sp. |
|
Topi3 |
Orange microcolonies and creamy in RC medium. Bacilli G (-), no esporulated. |
Aerobic, mobile, glucose (-), lactose (-). |
Pseudomonas sihuiensis |
|
Topi5 |
Red microcolonies and creamy in RC medium. Bacilli G (-), no esporulated. |
Aerobic, mobile, glucose (-), lactose (-). |
- |
|
Topi9 |
Red microcolonies and creamy in RC medium. Bacilli G (-), no esporulated. |
Aerobic, mobile, glucose (-), lactose (-). |
- |
|
Topi10 |
Red microcolonies and creamy in RC medium. Bacilli G (-), no esporulated. |
Aerobic, mobile, glucose (-), lactose (-). |
- |
|
Yacón |
Ya3 |
Red microcolonies and consistent in RC medium. Bacilli G (-), no esporulated. |
Aerobic, mobile, glucose (-), lactose (-). |
Pseudomonas alcaligenes |
Ya6 |
Red microcolonies and creamy in RC medium. Bacilli G (-), no esporulated. |
Aerobic, mobile, glucose (-), lactose (-). |
- |
|
Ya11 |
Red microcolonies and fluid in RC medium. Bacilli G (-), no esporulated. |
Aerobic, mobile, glucose (-), lactose (-). |
Pseudomonas resinovorans |
|
Ya15 |
Red microcolonies and creamy in RC medium. Bacilli G (-), no esporulated. |
Aerobic, mobile, glucose (-), lactose (-). |
- |
|
YaMi |
Red microcolonies and creamy in RC medium. Bacilli G (-), no esporulated. |
Aerobic, mobile, glucose (-), lactose (-). |
Pseudomonas sihuiensis |
Table 1 Molecular characterization and identification of the different isolates of N-fixing bacteria obtained from the endorhizosphere of topinambur and yacón cultures
Analysis of the 16S rRNA sequence determined that the molecularly identified bacteria belong to the genus Pseudomonas. Of the three isolates from the topinambur endorhizosphere, two of them corresponded to the P. sihuiensis species and the rest to Pseudomonas sp. In reference to the isolates obtained from yacón plants, they corresponded to the species Pseudomonas alcaligenes, P. resinovorans and P. sihuiensis (Table 1) (Figure 2).
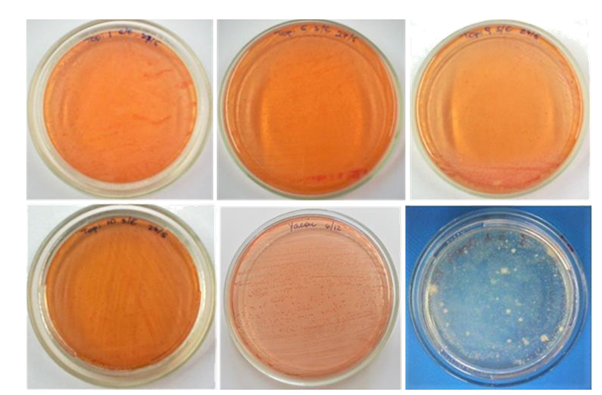
Figure 2 Colonies of nitrogen-free fixing bacteria from the endorhizosphere of topinambur (Helianthus tuberosus) and yacón (Smallanthus sonchifolius).
Pseudomonas It is a genus of gram-negative aerobic gammaproteobacteria, belonging to the Pseudomonadaceae family that contains 191 described species.39 Members of the genus demonstrate great metabolic diversity and, consequently, are capable of colonizing a wide range of niches.40
Different species of the genus Pseudomonas are used in the manufacture of biofertizers not only as FBN, but also as phosphorus solubilizers, PGPR, and mobilizers of zinc and potassium.6 However, the molecularly identified isolates correspond to microorganisms reported in other countries and studied in the bioremediation of soils and waters contaminated by their ability to biodegrade hydrocarbons, pesticides and certain chemical substances,41–44 but without agricultural relevance. Such as, P. sihuiensis was isolated from forest soils of southern China45 and in Brazil in sites with a history of frequent oil spills46 and studied for its ability to biodegrade hydrocarbons in seawater and its application in the bioremediation of marine environments due to oil spills.46 Furthermore, P. alcaligenes is used in antibacterial tests with the aim of evaluating the antimicrobial properties of oils essential substances,47 antagonistic activity against plant pathogens.48 While, P. resinovorans studied to know gene transcription mechanisms and enzymatic activity,41,49,50 it can be a human pathogen, but the cases are very rare (Figure 3).51,52
The investigation indicated the presence of bacteria of the Pseudomonas genus as free fixatives of atmospheric nitrogen in the endorhizosphere of topinambur and yacon grown in soils of the Central Valley of the Province of Catamarca. The species Pseudomonas sihuiensis, P. alcaligenes and P. resinovorans are reported for the first time in the province of Catamarca.
None.
Authors declare no conflict of interestexists.

©2020 Gabriela, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.